Rpd3S组蛋白去乙酰化酶复合物核小体去乙酰化和DNA连接子紧固的结构基础。
IF 28.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
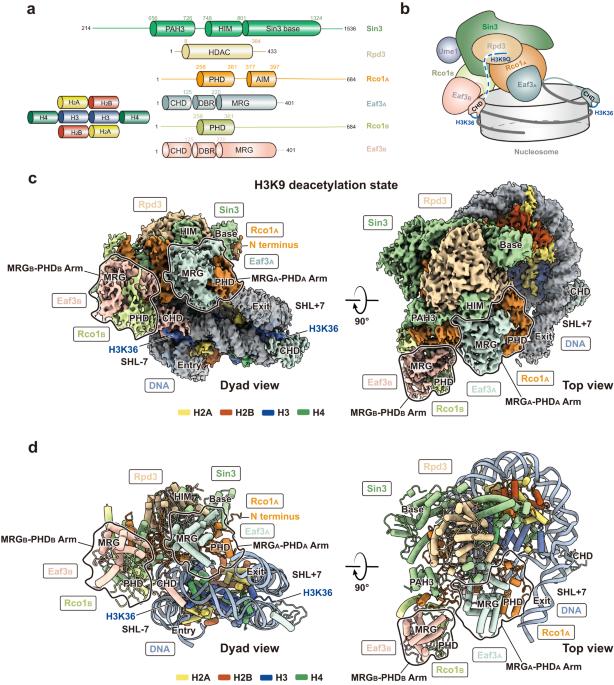
在酿酒酵母中,Sin3组蛋白脱乙酰酶(HDAC)复合物Rpd3S的活性阻止了编码区的隐性转录,该复合物由转录RNA聚合酶II(RNAPII)携带以脱乙酰和稳定染色质。尽管Rpd3S具有根本的重要性,但其脱乙酰核小体和调节染色质动力学的机制仍然难以捉摸。在这里,我们确定了Rpd3S与核小体核心颗粒(NCPs)复合物的几种冷冻EM结构,包括H3/H4脱乙酰状态、替代脱乙酰状态,连接体收紧状态,以及Rpd3S和NCPs上的Hho1连接体组蛋白共存的状态。这些结构表明,Rpd3S利用保守的Sin3碱性表面,在其与H3K36甲基化和核小体外DNA连接子的相互作用的指导下,在核小体DNA中导航,以靶向乙酰化的H3K9并对其他组蛋白尾部进行采样。此外,我们的结构表明,Rpd3S重新配置DNA连接子,并与Hho1协同作用以参与NCP,这可能揭示了Rpd3S和Hho1如何在基因沉默中协同工作。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex
In Saccharomyces cerevisiae, cryptic transcription at the coding region is prevented by the activity of Sin3 histone deacetylase (HDAC) complex Rpd3S, which is carried by the transcribing RNA polymerase II (RNAPII) to deacetylate and stabilize chromatin. Despite its fundamental importance, the mechanisms by which Rpd3S deacetylates nucleosomes and regulates chromatin dynamics remain elusive. Here, we determined several cryo-EM structures of Rpd3S in complex with nucleosome core particles (NCPs), including the H3/H4 deacetylation states, the alternative deacetylation state, the linker tightening state, and a state in which Rpd3S co-exists with the Hho1 linker histone on NCP. These structures suggest that Rpd3S utilizes a conserved Sin3 basic surface to navigate through the nucleosomal DNA, guided by its interactions with H3K36 methylation and the extra-nucleosomal DNA linkers, to target acetylated H3K9 and sample other histone tails. Furthermore, our structures illustrate that Rpd3S reconfigures the DNA linkers and acts in concert with Hho1 to engage the NCP, potentially unraveling how Rpd3S and Hho1 work in tandem for gene silencing.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


