埃塞俄比亚出现了对青蒿素具有耐药性的恶性疟原虫和诊断方法。
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
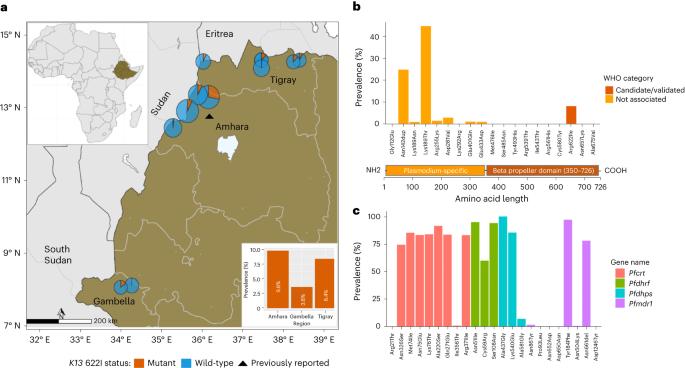
诊断和治疗恶性疟原虫感染是有效控制疟疾的必要条件,也是消灭疟疾工作的先决条件;因此,我们需要监测耐药性和诊断耐药性寄生虫的出现、进化和传播。我们对埃塞俄比亚一项诊断耐药性监测研究中收集的609例疟疾病例的寄生虫基因组中的关键耐药性突变和1832个SNPs进行了深度测序。我们发现8.0%(95%CI 7.0-9.0)的疟疾病例是由携带候选青蒿素部分耐药性kelch13(K13)622I突变的恶性疟原虫引起的,该突变在由富含组氨酸的蛋白2和3(pfhrp2/3)缺失介导的诊断抗性寄生虫中比在野生型寄生虫中更不常见(P = 亲缘关系分析表明,K13 622I寄生虫之间的亲缘关系显著高于野生型(P本文章由计算机程序翻译,如有差异,请以英文原文为准。
Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia
Diagnosis and treatment of Plasmodium falciparum infections are required for effective malaria control and are pre-requisites for malaria elimination efforts; hence we need to monitor emergence, evolution and spread of drug- and diagnostics-resistant parasites. We deep sequenced key drug-resistance mutations and 1,832 SNPs in the parasite genomes of 609 malaria cases collected during a diagnostic-resistance surveillance study in Ethiopia. We found that 8.0% (95% CI 7.0–9.0) of malaria cases were caused by P. falciparum carrying the candidate artemisinin partial-resistance kelch13 (K13) 622I mutation, which was less common in diagnostic-resistant parasites mediated by histidine-rich proteins 2 and 3 (pfhrp2/3) deletions than in wild-type parasites (P = 0.03). Identity-by-descent analyses showed that K13 622I parasites were significantly more related to each other than to wild type (P < 0.001), consistent with recent expansion and spread of this mutation. Pfhrp2/3-deleted parasites were also highly related, with evidence of clonal transmissions at the district level. Of concern, 8.2% of K13 622I parasites also carried the pfhrp2/3 deletions. Close monitoring of the spread of combined drug- and diagnostic-resistant parasites is needed. Plasmodium falciparum candidate artemisinin partial-resistance Kelch13 622I mutation co-occurs with pfhrp2/3 deletions in Ethiopia.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


