利用ISSR和SSR标记对伊朗面包小麦野生近缘的遗传多样性进行研究。
IF 3.5
Q3 Biochemistry, Genetics and Molecular Biology
Journal of Genetic Engineering and Biotechnology
Pub Date : 2023-12-01
DOI:10.1186/s43141-023-00526-5
引用次数: 0
摘要
背景:小麦野生近缘系是小麦育种中最重要的遗传资源之一。因此,确定小麦的野生近缘并认识其多样性,无疑是扩大基因库的丰富性和新品种遗传基础的有效手段,也是今后育种工作的一个有用工具。利用SSR和ISSR两种dna标记对伊朗国家植物基因库中49个稻瘟病属(Aegilops)和小麦属(Triticum)的分子多样性进行了分析。此外,本研究还旨在探讨不同遗传背景的材料之间的关系。结果:10条SSR引物和1条ISSR引物分别产生2065条和1524条多态性条带。SSR标记的多态性带数(NPB)、多态性信息含量(PIC)、标记指数(MI)和分辨能力(Rp)分别为162 ~ 317、0.830 ~ 0.919、1.326 ~ 3.167和3.169 ~ 5.692,ISSR标记的多态性带数(NPB)、多态性信息含量(PIC)和分辨能力(Rp)分别为103 ~ 185、0.377 ~ 0.441、0.660 ~ 1.151和3.169 ~ 5.693。这表明这两个标记在检测所研究的材料的多态性方面是有效的。ISSR标记的多态性率、MI和Rp均高于SSR标记。两种标记的分子变异分析表明,种内遗传变异大于种间遗传多样性。在小麦和小麦中发现的高水平的基因组多样性为发现小麦育种有用的基因提供了理想的基因库。采用UPGMA聚类分析方法,根据SSR和ISSR标记将材料分为8个类群。聚类分析结果表明,在多数情况下,尽管同一省份的植物遗传资源具有相似性,但其地理格局与分子聚类分析结果并不一致。根据坐标分析,相邻群体的相似性最大,而距离较远的群体之间的遗传距离最大。遗传结构分析成功地分离了材料的倍性水平。结论:这两个标记提供了一种完整的伊朗小麦属和小麦属遗传多样性模型。本研究使用的引物有效、信息量大、具有基因组特异性,可用于基因组解释实验。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Investigation of genetic diversity of Iranian wild relatives of bread wheat using ISSR and SSR markers
Background
Wild relatives of wheat are one of the most important genetic resources to use in wheat breeding programs. Therefore, identifying wild relatives of wheat and being aware of their diversity, is undeniably effective in expanding the richness of the gene pool and the genetic base of new cultivars and can be a useful tool for breeders in the future. The present study was performed to evaluate the molecular diversity among 49 accessions of the genera Aegilops and Triticum in the National Plant Gene Bank of Iran using two DNA-based markers, i.e., SSR and ISSR. Also, the present study aimed to examine the relationships among the accessions studied belonging to different genetic backgrounds.
Results
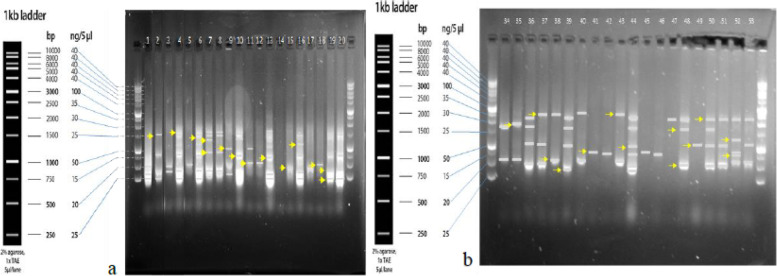
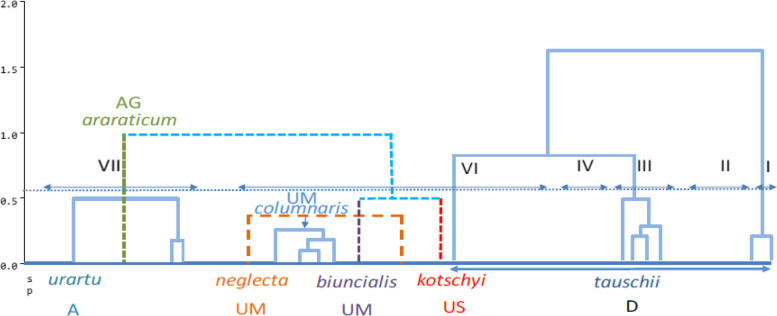
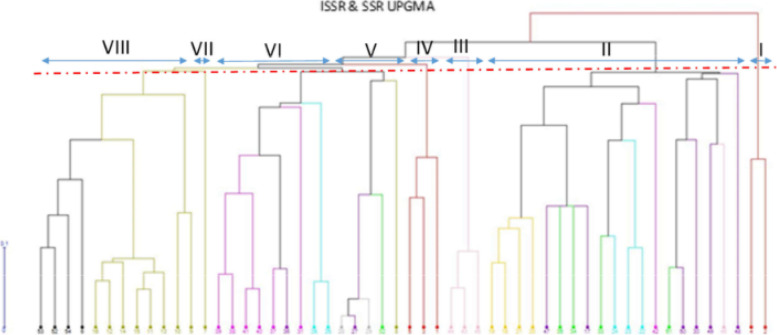
Ten SSR and tan ISSR primers produced 2065 and 1524 polymorphism bands, respectively. The number of Polymorphic Bands (NPB), the Polymorphism Information Content (PIC), Marker Index (MI), and Resolving Power (Rp) in SSR marker was 162 to 317, 0.830 to 0.919, 1.326 to 3.167, and 3.169 to 5.692, respectively, and in the ISSR marker, it was from 103 to 185, 0.377 to 0.441, 0.660 to 1.151, and 3.169 to 5.693, respectively. This indicates the efficiency of both markers in detecting polymorphism among the accessions studied. The ISSR marker had a higher polymorphism rate, MI, and Rp than the SSR marker. Molecular analysis of variance for both DNA-based markers showed that the genetic variation within the species was more than the genetic diversity between them. The high level of genomic diversity discovered in the Aegilops and Triticum species proved to provide an ideal gene pool for discovering genes useful for wheat breeding. The accessions were classified into eight groups based on SSR and ISSR markers using the UPGMA method of cluster analysis. According to the cluster analysis results, despite similarities between the accessions of a given province, in most cases, the geographical pattern was not in accordance with that observed using the molecular clustering. Based on the coordinate analysis, neighboring groups showed the maximum similarities, and distant ones revealed the maximum genetic distance from each other. The genetic structure analysis successfully separated accessions for their ploidy levels.
Conclusions
Both markers provided a comprehensive model of genetic diversity between Iranian accessions of Aegilops and Triticum genera. Primers used in the present study were effective, informative, and genome-specific which could be used in genome explanatory experiments.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Genetic Engineering and Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
5.70
自引率
5.70%
发文量
159
审稿时长
16 weeks
期刊介绍:
Journal of genetic engineering and biotechnology is devoted to rapid publication of full-length research papers that leads to significant contribution in advancing knowledge in genetic engineering and biotechnology and provide novel perspectives in this research area. JGEB includes all major themes related to genetic engineering and recombinant DNA. The area of interest of JGEB includes but not restricted to: •Plant genetics •Animal genetics •Bacterial enzymes •Agricultural Biotechnology, •Biochemistry, •Biophysics, •Bioinformatics, •Environmental Biotechnology, •Industrial Biotechnology, •Microbial biotechnology, •Medical Biotechnology, •Bioenergy, Biosafety, •Biosecurity, •Bioethics, •GMOS, •Genomic, •Proteomic JGEB accepts
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


