酵母Sis1与Hsp70 c端EEVD肽复合物中残基1-81的主链和侧链NMR定位。
IF 0.8
4区 生物学
Q4 BIOPHYSICS
引用次数: 0
摘要
分子伴侣有助于蛋白质折叠和组装,而不改变其最终结构,在几个折叠过程中,需要Hsp70和Hsp40家族成员之间的相互作用。在这里,我们报道了来自酿酒酵母Sis1的B类Hsp40的J结构域的主链和侧链的1H、15N和13C核与Hsp70的C末端EEVD基序复合的NMR化学位移分配。这些数据揭示了关于结构和骨架动力学的信息,这些信息大大增加了对J结构域-Hsp70-EEVD相互作用机制的理解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
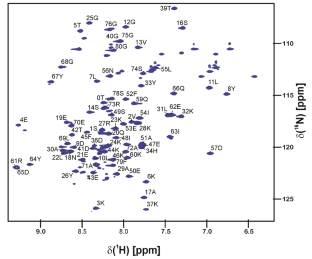
Backbone and sidechain NMR assignments of residues 1–81 from yeast Sis1 in complex with an Hsp70 C-terminal EEVD peptide
Molecular chaperones aid proteins to fold and assemble without modifying their final structure, requiring, in several folding processes, the interplay between members of the Hsp70 and Hsp40 families. Here, we report the NMR chemical shift assignments for 1 H, 15 N, and 13 C nuclei of the backbone and side chains of the J-domain of the class B Hsp40 from Saccharomyces cerevisiae, Sis1, complexed with the C-terminal EEVD motif of Hsp70. The data revealed information on the structure and backbone dynamics that add significantly to the understanding of the J-domain-Hsp70-EEVD mechanism of interaction.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Biomolecular NMR Assignments
生物-光谱学
CiteScore
1.70
自引率
11.10%
发文量
59
审稿时长
6-12 weeks
期刊介绍:
Biomolecular NMR Assignments provides a forum for publishing sequence-specific resonance assignments for proteins and nucleic acids as Assignment Notes. Chemical shifts for NMR-active nuclei in macromolecules contain detailed information on molecular conformation and properties.
Publication of resonance assignments in Biomolecular NMR Assignments ensures that these data are deposited into a public database at BioMagResBank (BMRB; http://www.bmrb.wisc.edu/), where they are available to other researchers. Coverage includes proteins and nucleic acids; Assignment Notes are processed for rapid online publication and are published in biannual online editions in June and December.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


