HSPA8 acts as an amyloidase to suppress necroptosis by inhibiting and reversing functional amyloid formation
IF 28.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
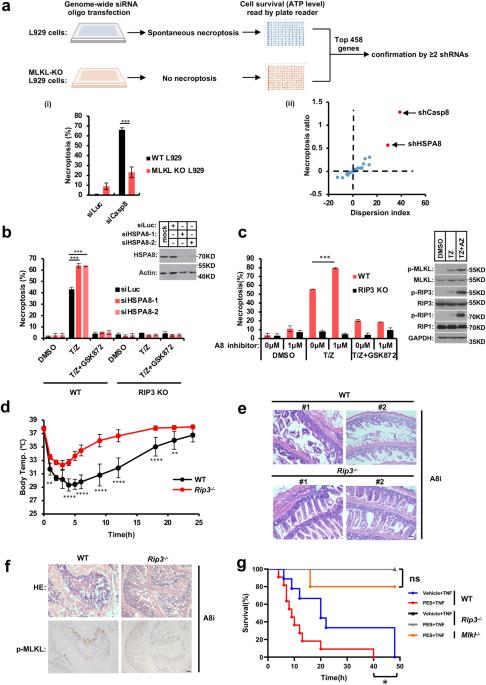
Ultra-stable fibrous structure is a hallmark of amyloids. In contrast to canonical disease-related amyloids, emerging research indicates that a significant number of cellular amyloids, termed ‘functional amyloids’, contribute to signal transduction as temporal signaling hubs in humans. However, it is unclear how these functional amyloids are effectively disassembled to terminate signal transduction. RHIM motif-containing amyloids, the largest functional amyloid family discovered thus far, play an important role in mediating necroptosis signal transduction in mammalian cells. Here, we identify heat shock protein family A member 8 (HSPA8) as a new type of enzyme — which we name as ‘amyloidase’ — that directly disassembles RHIM-amyloids to inhibit necroptosis signaling in cells and mice. Different from its role in chaperone-mediated autophagy where it selects substrates containing a KFERQ-like motif, HSPA8 specifically recognizes RHIM-containing proteins through a hydrophobic hexapeptide motif N(X1)φ(X3). The SBD domain of HSPA8 interacts with RHIM-containing proteins, preventing proximate RHIM monomers from stacking into functional fibrils; furthermore, with the NBD domain supplying energy via ATP hydrolysis, HSPA8 breaks down pre-formed RHIM-amyloids into non-functional monomers. Notably, HSPA8’s amyloidase activity in disassembling functional RHIM-amyloids does not require its co-chaperone system. Using this amyloidase activity, HSPA8 reverses the initiator RHIM-amyloids (formed by RIP1, ZBP1, and TRIF) to prevent necroptosis initiation, and reverses RIP3-amyloid to prevent necroptosis execution, thus eliminating multi-level RHIM-amyloids to effectively prevent spontaneous necroptosis activation. The discovery that HSPA8 acts as an amyloidase dismantling functional amyloids provides a fundamental understanding of the reversibility nature of functional amyloids, a property distinguishing them from disease-related amyloids that are unbreakable in vivo.
HSPA8作为淀粉样蛋白酶通过抑制和逆转功能性淀粉样蛋白的形成来抑制坏死。
超稳定的纤维结构是淀粉样蛋白的标志。与典型的疾病相关淀粉样蛋白相反,新兴的研究表明,大量的细胞淀粉样蛋白,称为“功能性淀粉样蛋白”,作为人类的时间信号中枢,有助于信号转导。然而,目前尚不清楚这些功能性淀粉样蛋白是如何被有效地分解以终止信号转导的。含有RHIM基序的淀粉样蛋白是迄今为止发现的最大的功能性淀粉样蛋白家族,在哺乳动物细胞中介导坏死信号转导中发挥着重要作用。在这里,我们确定热休克蛋白家族A成员8(HSPA8)是一种新型酶,我们称之为“淀粉样酶”,它直接分解RHIM淀粉样蛋白,以抑制细胞和小鼠的坏死信号。与在伴侣介导的自噬中选择含有KFERQ样基序的底物不同,HSPA8通过疏水性六肽基序N(X1)φ(X3)特异性识别含有RHIM的蛋白质。HSPA8的SBD结构域与含有RHIM的蛋白质相互作用,防止邻近的RHIM单体堆积成功能原纤维;此外,NBD结构域通过ATP水解提供能量,HSPA8将预先形成的RHIM淀粉样蛋白分解为非功能单体。值得注意的是,HSPA8在分解功能性RHIM淀粉样蛋白中的淀粉样酶活性不需要其共同伴侣系统。利用这种淀粉样酶活性,HSPA8逆转引发剂RHIM淀粉样蛋白(由RIP1、ZBP1和TRIF形成)以防止坏死起始,并逆转RIP3淀粉样蛋白以防止坏死执行,从而消除多级RHIM淀粉状蛋白以有效防止自发坏死激活。HSPA8作为一种淀粉样酶分解功能性淀粉样蛋白的发现,为功能性淀粉类蛋白的可逆性提供了基本的理解,这一特性将其与体内牢不可破的疾病相关淀粉样蛋白区分开来。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


