Concise Cascade Methods for Transgenic Rice Seed Discrimination using Spectral Phenotyping.
IF 6.4
1区 农林科学
Q1 AGRONOMY
引用次数: 0
Abstract
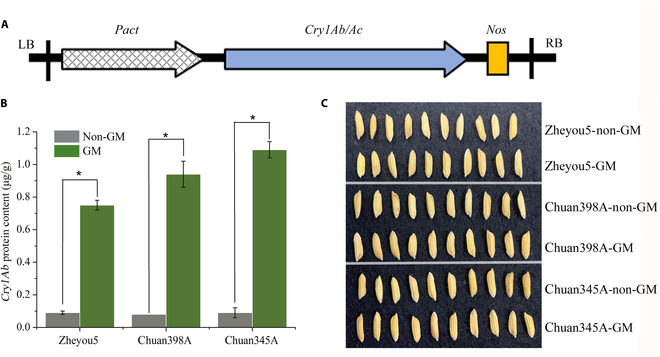
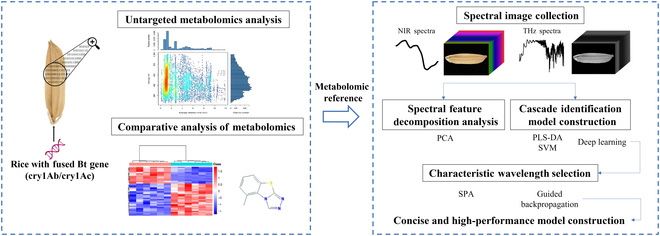
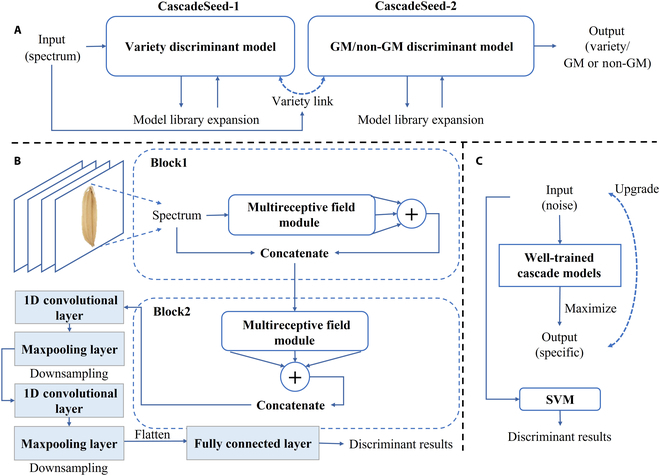
Currently, the presence of genetically modified (GM) organisms in agro-food markets is strictly regulated by enacted legislation worldwide. It is essential to ensure the traceability of these transgenic products for food safety, consumer choice, environmental monitoring, market integrity, and scientific research. However, detecting the existence of GM organisms involves a combination of complex, time-consuming, and labor-intensive techniques requiring high-level professional skills. In this paper, a concise and rapid pipeline method to identify transgenic rice seeds was proposed on the basis of spectral imaging technologies and the deep learning approach. The composition of metabolome across 3 rice seed lines containing the cry1Ab/cry1Ac gene was compared and studied, substantiating the intrinsic variability induced by these GM traits. Results showed that near-infrared and terahertz spectra from different genotypes could reveal the regularity of GM metabolic variation. The established cascade deep learning model divided GM discrimination into 2 phases including variety classification and GM status identification. It could be found that terahertz absorption spectra contained more valuable features and achieved the highest accuracy of 97.04% for variety classification and 99.71% for GM status identification. Moreover, a modified guided backpropagation algorithm was proposed to select the task-specific characteristic wavelengths for further reducing the redundancy of the original spectra. The experimental validation of the cascade discriminant method in conjunction with spectroscopy confirmed its viability, simplicity, and effectiveness as a valuable tool for the detection of GM rice seeds. This approach also demonstrated its great potential in distilling crucial features for expedited transgenic risk assessment.
利用光谱表型鉴别转基因水稻种子的简明级联方法。
目前,转基因生物在农产品市场上的存在受到世界各国立法的严格管制。确保这些转基因产品的可追溯性对于食品安全、消费者选择、环境监测、市场诚信和科学研究至关重要。然而,检测转基因生物的存在涉及复杂、耗时和劳动密集型技术的结合,需要高水平的专业技能。本文基于光谱成像技术和深度学习方法,提出了一种简洁、快速的转基因水稻种子管道识别方法。对含cry1Ab/cry1Ac基因的3个水稻种子品系的代谢组组成进行了比较研究,证实了这些转基因性状诱导的内在变异。结果表明,不同基因型的近红外和太赫兹光谱可以揭示转基因代谢变异的规律。建立的级联深度学习模型将转基因识别分为品种分类和转基因状态识别两个阶段。结果表明,太赫兹吸收光谱包含更多有价值的特征,对品种分类和转基因状态鉴定的准确率最高,分别为97.04%和99.71%。此外,提出了一种改进的制导反向传播算法来选择特定任务的特征波长,以进一步减少原始光谱的冗余。实验验证了级联判别法与光谱法的结合,证实了其作为一种有价值的转基因水稻种子检测工具的可行性、简单性和有效性。该方法还显示了其在提取关键特征以加快转基因风险评估方面的巨大潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Plant Phenomics
Multiple-
CiteScore
8.60
自引率
9.20%
发文量
26
审稿时长
14 weeks
期刊介绍:
Plant Phenomics is an Open Access journal published in affiliation with the State Key Laboratory of Crop Genetics & Germplasm Enhancement, Nanjing Agricultural University (NAU) and published by the American Association for the Advancement of Science (AAAS). Like all partners participating in the Science Partner Journal program, Plant Phenomics is editorially independent from the Science family of journals.
The mission of Plant Phenomics is to publish novel research that will advance all aspects of plant phenotyping from the cell to the plant population levels using innovative combinations of sensor systems and data analytics. Plant Phenomics aims also to connect phenomics to other science domains, such as genomics, genetics, physiology, molecular biology, bioinformatics, statistics, mathematics, and computer sciences. Plant Phenomics should thus contribute to advance plant sciences and agriculture/forestry/horticulture by addressing key scientific challenges in the area of plant phenomics.
The scope of the journal covers the latest technologies in plant phenotyping for data acquisition, data management, data interpretation, modeling, and their practical applications for crop cultivation, plant breeding, forestry, horticulture, ecology, and other plant-related domains.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


