A computational study on mitogenome-encoded proteins of Pavo cristatus and Pavo muticus identifies key genetic variations with functional implications
IF 3.5
Q3 Biochemistry, Genetics and Molecular Biology
Journal of Genetic Engineering and Biotechnology
Pub Date : 2023-12-01
DOI:10.1186/s43141-023-00534-5
引用次数: 0
Abstract
Background
The Pavo cristatus population, native to the Indian subcontinent, is thriving well in India. However, the Pavo muticus population, native to the tropical forests of Southeast Asia, has reduced drastically and has been categorised as an endangered group. To understand the probable genetic factors associated with the decline of P. muticus, we compared the mitogenome-encoded proteins (13 proteins) between these two species.
Results
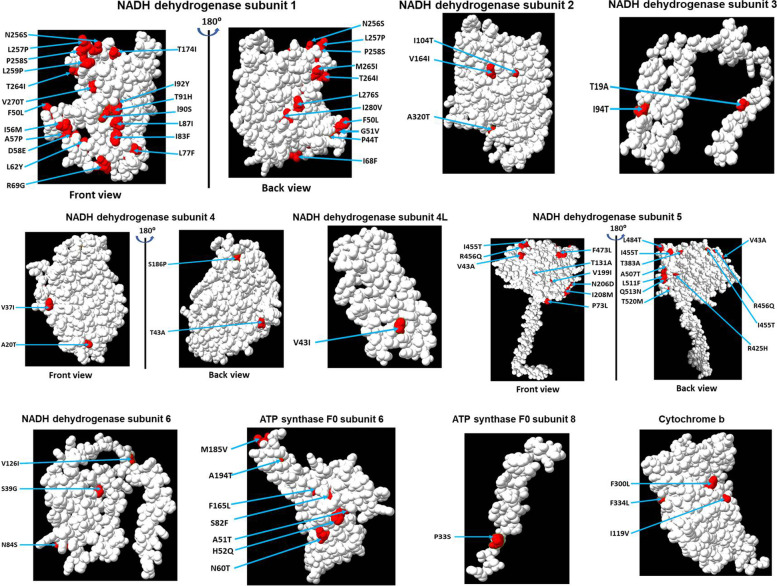
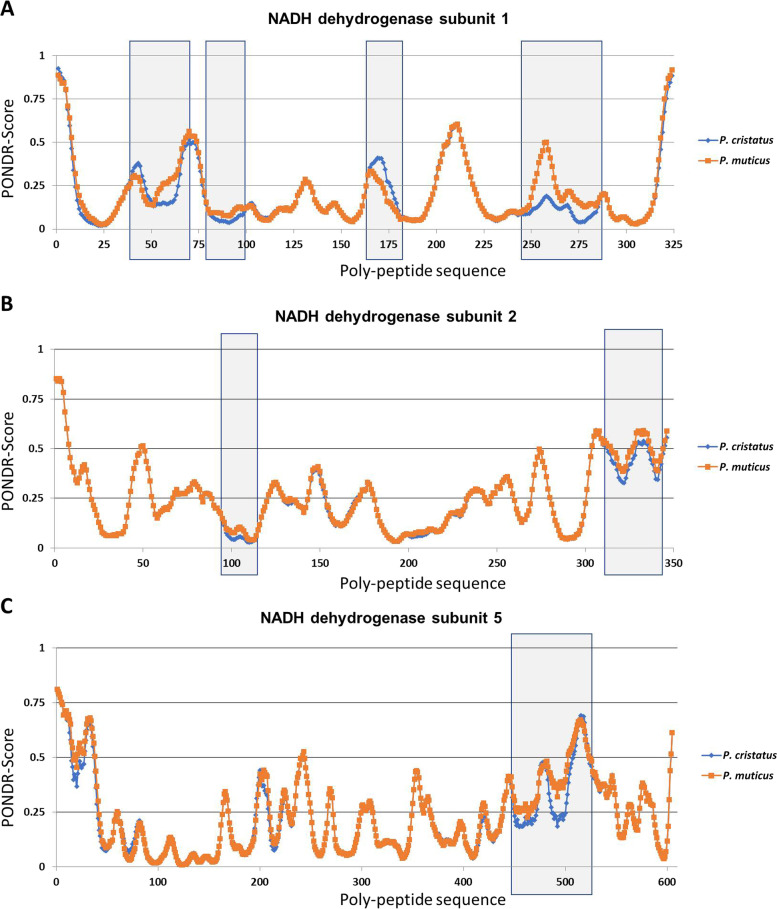
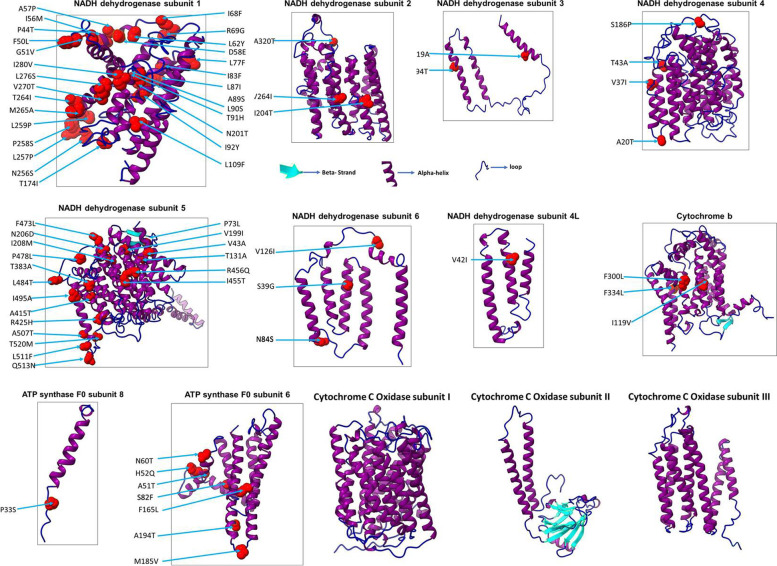
Our data revealed that the most frequent variant between these two species was mtND1, which had an alteration in 9.57% residues, followed by mtND5 and mtATP6. We extended our study on the rest of the proteins and observed that cytochrome c oxidase subunits 1, 2, and 3 do not have any change. The 3-dimensional structure of all 13 proteins was modeled using the Phyre2 programme. Our data show that most of the proteins are alpha helical, and the variations observed in P. muticus reside on the surface of the respective proteins. The effect of variation on protein function was also predicted, and our results show that amino acid substitution in mtND1 at 14 sites could be deleterious. Similarly, destabilising changes were observed in mtND1, 2, 3, 4, 5, and 6 and mtATP6–8 due to amino acid substitution in P. muticus. Furthermore, protein disorder scores were considerably altered in mtND1, 2, and 5 of P. muticus.
Conclusions
The results presented here strongly suggest that variations in mitogenome-encoded proteins of P. cristatus and P. muticus may alter their structure and functions. Subsequently, these variations could alter energy production and may correlate with the decline in the population of P. muticus.
一个计算研究Pavo cristatus和Pavo muticus的有丝分裂基因组编码蛋白识别关键的遗传变异与功能的影响。
背景:原产于印度次大陆的Pavo cristatus种群在印度蓬勃发展。然而,原产于东南亚热带森林的Pavo muticus种群数量急剧减少,已被列为濒危物种。为了了解与变异假单胞虫数量下降相关的可能遗传因素,我们比较了这两个物种的有丝分裂基因组编码蛋白(13种蛋白)。结果:我们的数据显示,这两个物种之间最常见的变异是mtND1,有9.57%的残基改变,其次是mtND5和mtATP6。我们扩展了对其余蛋白质的研究,观察到细胞色素c氧化酶亚基1、2和3没有任何变化。所有13种蛋白的三维结构都使用Phyre2程序建模。我们的数据表明,大多数蛋白质是α螺旋状的,在p.m uticus中观察到的变异存在于各自蛋白质的表面。我们还预测了变异对蛋白质功能的影响,结果表明mtND1中14个位点的氨基酸取代可能是有害的。同样,在p.a muticus中,由于氨基酸替换,mtND1、2、3、4、5、6和mtATP6-8也发生了不稳定变化。此外,muticus的mtND1、2和5蛋白紊乱评分显著改变。结论:本研究结果强烈提示cristatus和muticus有丝分裂基因组编码蛋白的变异可能改变了它们的结构和功能。随后,这些变化可能会改变能量的产生,并可能与木斑蝽种群的减少有关。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Genetic Engineering and Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
5.70
自引率
5.70%
发文量
159
审稿时长
16 weeks
期刊介绍:
Journal of genetic engineering and biotechnology is devoted to rapid publication of full-length research papers that leads to significant contribution in advancing knowledge in genetic engineering and biotechnology and provide novel perspectives in this research area. JGEB includes all major themes related to genetic engineering and recombinant DNA. The area of interest of JGEB includes but not restricted to: •Plant genetics •Animal genetics •Bacterial enzymes •Agricultural Biotechnology, •Biochemistry, •Biophysics, •Bioinformatics, •Environmental Biotechnology, •Industrial Biotechnology, •Microbial biotechnology, •Medical Biotechnology, •Bioenergy, Biosafety, •Biosecurity, •Bioethics, •GMOS, •Genomic, •Proteomic JGEB accepts
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


