Molecular identification of Anaplasma platys in cattle by nested PCR.
IF 1.3
Q4 MICROBIOLOGY
引用次数: 0
Abstract
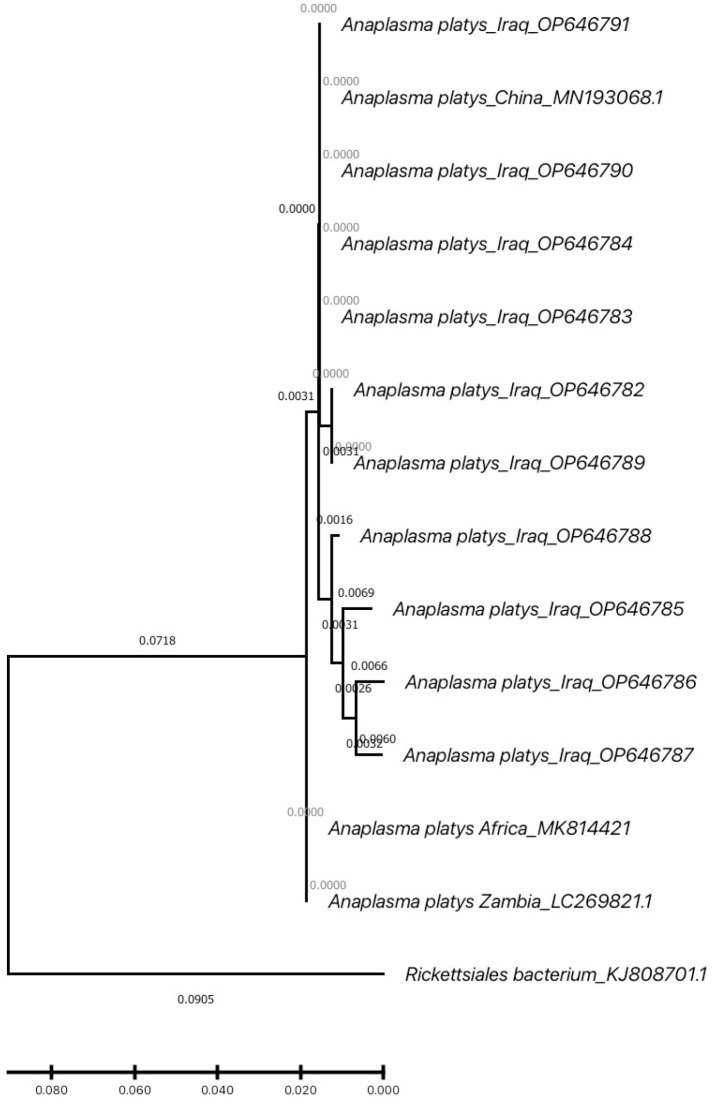
Background and Objectives: Anaplasmosis is a zoonotic disease caused by Gram-negative bacterium from Anaplasmataceae family. Anaplasma causes high economic losses worldwide. 16S rRNA analysis was used to diagnose Anaplasma platys in Cattle. Phylogenetic tree and estimation of evolutionary divergence between A. platys isolates were performed. Materials and Methods: A total of 60 blood samples were collected from a cattle farm in AL-Diwaniyah province. 16S rRNA gene was identified using nested PCR. Overall, 40% of cattle that were chosen to collect the blood were identified to be infected with A. platys. Results: The results have shown presence of targeting partial region of 16S rRNA gene in 24 samples out of 60. Sequencing results of 10 samples have revealed that the phylogenetic tree was divided in to two separate clades. Five isolates of A. platys-Iraq (accession no. OP646782, OP646783, OP646784, OP646790, and OP646791) were located in one clade with the A. platys-China (accession no. MN193068.1). While, five isolates (accession no. OP646785, OP646786, OP646787, OP646788, OP646789) were in different clade with two isolates of A. platys-Africa and A. platys-Zambia in distinct branches, close to the Rickettsiales. Conclusion: The phylogenetic study of A. platys sequences indicated that the isolates were collected from a cattle farm in Al-Dewaniyah were similar and close related to A. platys-China, A. platys-Zambia and A. platys-Africa). This study suggests that cattle can be considered a reservoir of A. platys.

用巢式PCR鉴定牛羊无原体的分子结构。
背景与目的:无形体病是由无形体科革兰氏阴性菌引起的人畜共患疾病。无形体在世界范围内造成巨大的经济损失。采用16S rRNA分析方法诊断牛无形体病。进行了系统发育树分析,并对不同分离株的进化差异进行了估计。材料和方法:从AL-Diwaniyah省的一个养牛场共采集了60份血液样本。采用巢式PCR法鉴定16S rRNA基因。总的来说,挑选采集血液的牛中有40%被鉴定为感染了鸭绒单胞虫。结果:60份样品中有24份存在16S rRNA基因的部分靶向区域。10个样本的测序结果显示,系统发育树被划分为两个独立的分支。5株伊拉克白斑绦虫分离株(加入号:OP646782, OP646783, OP646784, OP646790,和OP646791)与A. platys-China(加入号:MN193068.1)。5个分离株(accession no. 5)。OP646785、OP646786、OP646787、OP646788、OP646789)属于不同的进化支,非洲和赞比亚两个分离株在不同分支,与立克次体相近。结论:从Al-Dewaniyah的一个养牛场采集到的鸭胸蚜序列系统发育研究表明,该分离株与鸭胸蚜(中国)、鸭胸蚜(赞比亚)和鸭胸蚜(非洲)相似且关系密切。这项研究表明,牛可以被认为是一个储存库。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Iranian Journal of Microbiology
MICROBIOLOGY-
CiteScore
2.40
自引率
7.10%
发文量
96
审稿时长
12 weeks
期刊介绍:
The Iranian Journal of Microbiology (IJM) is an international, multi-disciplinary, peer-reviewed journal that provides rapid publication of the most advanced scientific research in the areas of basic and applied research on bacteria and other micro-organisms, including bacteria, viruses, yeasts, fungi, microalgae, and protozoa concerning the development of tools for diagnosis and disease control, epidemiology, antimicrobial agents, clinical microbiology, immunology, Genetics, Genomics and Molecular Biology. Contributions may be in the form of original research papers, review articles, short communications, case reports, technical reports, and letters to the Editor. Research findings must be novel and the original data must be available for review by the Editors, if necessary. Studies that are preliminary, of weak originality or merely descriptive as well as negative results are not appropriate for the journal. Papers considered for publication must be unpublished work (except in an abstract form) that is not under consideration for publication anywhere else, and all co-authors should have agreed to the submission. Manuscripts should be written in English.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


