Engineering of increased L-Threonine production in bacteria by combinatorial cloning and machine learning
Abstract
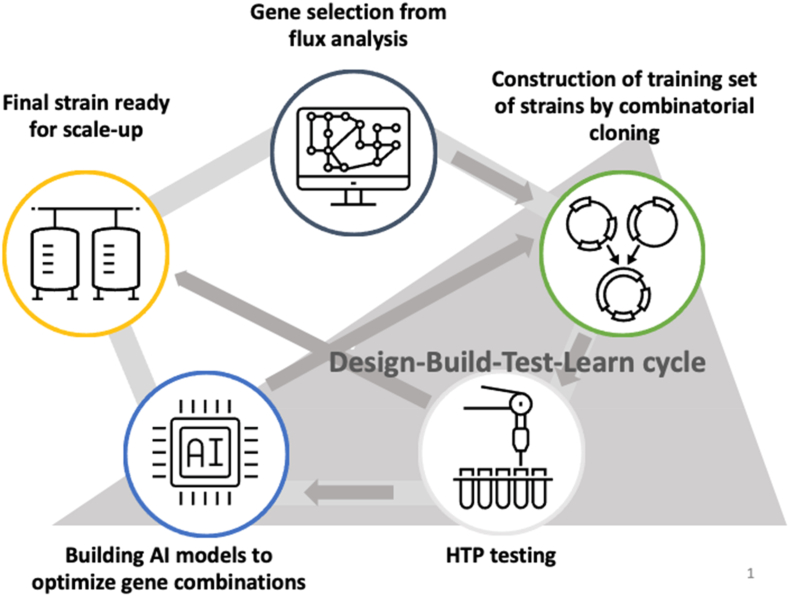
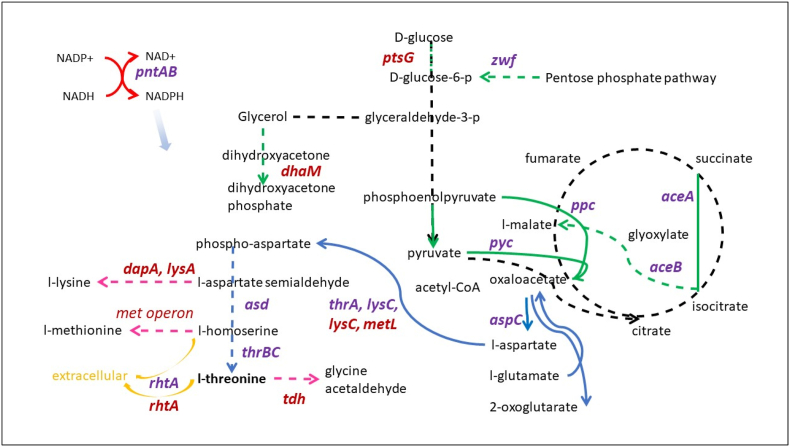
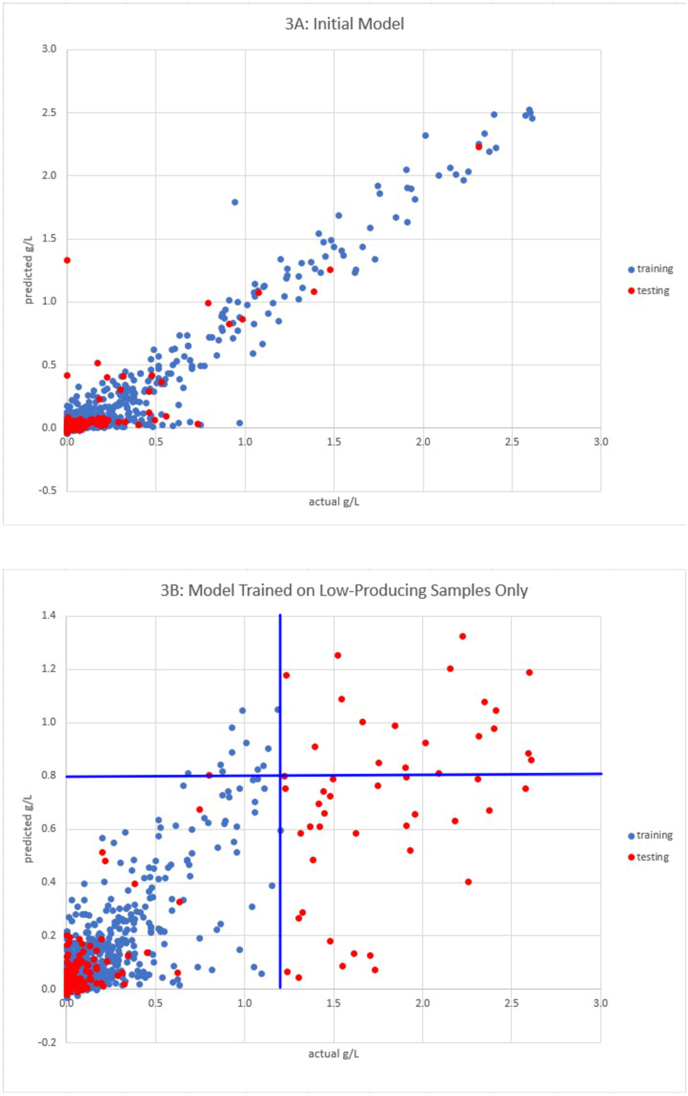
The goal of this study is to develop a general strategy for bacterial engineering using an integrated synthetic biology and machine learning (ML) approach. This strategy was developed in the context of increasing L-threonine production in Escherichia coli ATCC 21277. A set of 16 genes was initially selected based on metabolic pathway relevance to threonine biosynthesis and used for combinatorial cloning to construct a set of 385 strains to generate training data (i.e., a range of L-threonine titers linked to each of the specific gene combinations). Hybrid (regression/classification) deep learning (DL) models were developed and used to predict additional gene combinations in subsequent rounds of combinatorial cloning for increased L-threonine production based on the training data. As a result, E. coli strains built after just three rounds of iterative combinatorial cloning and model prediction generated higher L-threonine titers (from 2.7 g/L to 8.4 g/L) than those of patented L-threonine strains being used as controls (4–5 g/L). Interesting combinations of genes in L-threonine production included deletions of the tdh, metL, dapA, and dhaM genes as well as overexpression of the pntAB, ppc, and aspC genes. Mechanistic analysis of the metabolic system constraints for the best performing constructs offers ways to improve the models by adjusting weights for specific gene combinations. Graph theory analysis of pairwise gene modifications and corresponding levels of L-threonine production also suggests additional rules that can be incorporated into future ML models.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


