What is the “modified” CTAB protocol? Characterizing modifications to the CTAB DNA extraction protocol
IF 2.7
3区 生物学
Q2 PLANT SCIENCES
引用次数: 7
Abstract
Abstract Cetyltrimethylammonium bromide (CTAB)–based methods are widely used to isolate DNA from plant tissues, but the unique chemical composition of secondary metabolites among plant species has necessitated optimization. Research articles often cite a “modified” CTAB protocol without explicitly stating how the protocol had been altered, creating non‐reproducible studies. Furthermore, the various modifications that have been applied to the CTAB protocol have not been rigorously reviewed and doing so could reveal optimization strategies across study systems. We surveyed the literature for modified CTAB protocols used for the isolation of plant DNA. We found that every stage of the CTAB protocol has been modified, and we summarized those modifications to provide recommendations for extraction optimization. Future genomic studies will rely on optimized CTAB protocols. Our review of the modifications that have been used, as well as the protocols we provide here, could better standardize DNA extractions, allowing for repeatable and transparent studies.
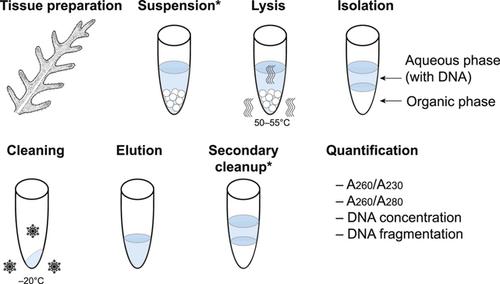
什么是“修改”的CTAB协议?描述对CTAB DNA提取方案的修改
基于十六烷基三甲基溴化铵(CTAB)的方法被广泛用于植物组织DNA的分离,但由于植物次生代谢物的化学成分独特,需要对其进行优化。研究文章经常引用“修改过的”CTAB协议,而没有明确说明该协议是如何被修改的,这造成了不可重复的研究。此外,应用于CTAB协议的各种修改尚未经过严格审查,这样做可以揭示跨研究系统的优化策略。我们查阅了用于分离植物DNA的改良CTAB协议的文献。我们发现CTAB协议的每个阶段都被修改了,我们总结了这些修改,为提取优化提供建议。未来的基因组研究将依赖于优化的CTAB协议。我们对已经使用的修改的回顾,以及我们在这里提供的协议,可以更好地标准化DNA提取,允许可重复和透明的研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Applications in Plant Sciences
PLANT SCIENCES-
CiteScore
7.30
自引率
0.00%
发文量
50
审稿时长
12 weeks
期刊介绍:
Applications in Plant Sciences (APPS) is a monthly, peer-reviewed, open access journal promoting the rapid dissemination of newly developed, innovative tools and protocols in all areas of the plant sciences, including genetics, structure, function, development, evolution, systematics, and ecology. Given the rapid progress today in technology and its application in the plant sciences, the goal of APPS is to foster communication within the plant science community to advance scientific research. APPS is a publication of the Botanical Society of America, originating in 2009 as the American Journal of Botany''s online-only section, AJB Primer Notes & Protocols in the Plant Sciences.
APPS publishes the following types of articles: (1) Protocol Notes describe new methods and technological advancements; (2) Genomic Resources Articles characterize the development and demonstrate the usefulness of newly developed genomic resources, including transcriptomes; (3) Software Notes detail new software applications; (4) Application Articles illustrate the application of a new protocol, method, or software application within the context of a larger study; (5) Review Articles evaluate available techniques, methods, or protocols; (6) Primer Notes report novel genetic markers with evidence of wide applicability.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


