Development of a metabolomics-based data analysis approach for identifying drug metabolites based on high-resolution mass spectrometry.
IF 3
3区 农林科学
Q2 FOOD SCIENCE & TECHNOLOGY
引用次数: 1
Abstract
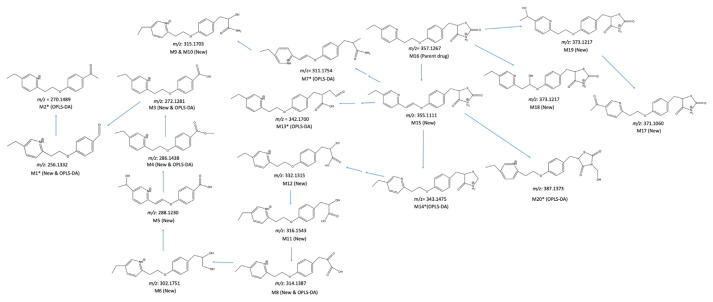
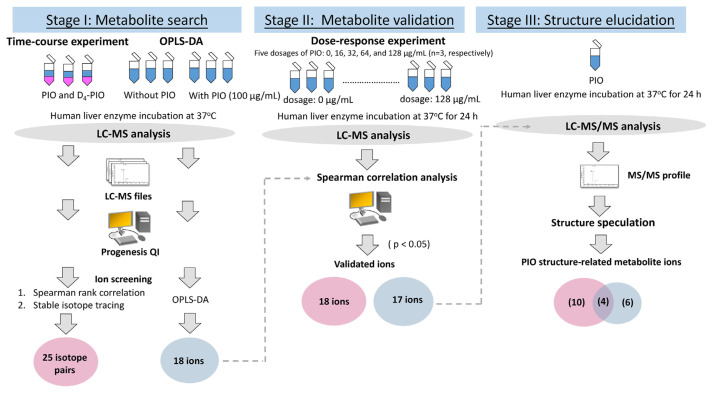
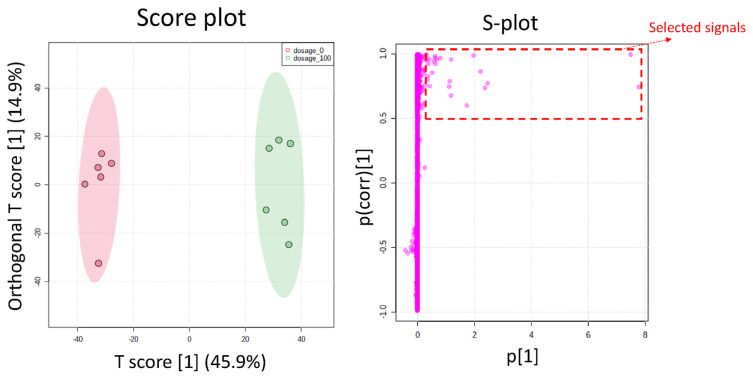
A metabolomics-based approach to data analysis is required for drug metabolites to be identified quickly. This study developed such an approach based on high-resolution mass spectrometry. Our approach is a two-stage one that combines a time-course experiment with stable isotope tracing. Pioglitazone (PIO) was used to improve glycemic management for type 2 diabetes mellitus. Consequently, PIO was taken as a model drug for identifying metabolites. During Stage I of data analysis, 704 out of 26626 ions exhibited a positive relationship between ion abundance ratio and incubation time in a time-course experiment. During Stage II, 25 isotope pairs were identified among the 704 ions. Among these 25 ions, 18 exhibited a dose-response relationship. Finally, 14 of the 18 ions were verified to be PIO structure-related metabolite ions. Otherwise, orthogonal partial least squares-discriminant analysis (OPLS-DA) was adopted to mine PIO metabolite ions, and 10 PIO structure-related metabolite ions were identified. However, only four ions were identified by both our developed approach and OPLS-DA, indicating that differences in the designs of metabolomics-based approaches to data analysis can result in differences in which metabolites are identified. A total of 20 PIO structure-related metabolites were identified by our developed approach and OPLS-DA, and six metabolites were novel. The results demonstrated that our developed two-stage data analysis approach can be used to effectively mine data on PIO metabolite ions from a relatively complex matrix.
开发基于代谢组学的数据分析方法,用于基于高分辨率质谱法识别药物代谢物。
需要一种基于代谢组学的数据分析方法来快速识别药物代谢物。本研究开发了一种基于高分辨率质谱的方法。我们的方法是一种两阶段的方法,将时间过程实验与稳定同位素示踪相结合。吡格列酮(PIO)用于改善2型糖尿病的血糖管理。因此,PIO被作为鉴定代谢物的模型药物。在数据分析的第一阶段,在一个时间过程实验中,26626个离子中有704个离子丰度比与孵育时间呈正相关。在第二阶段,在704个离子中鉴定出25对同位素。在这25个离子中,有18个离子表现出剂量效应关系。最后,18个离子中有14个被证实是PIO结构相关的代谢物离子。采用正交偏最小二乘判别分析(OPLS-DA)挖掘PIO代谢产物离子,鉴定出10个与PIO结构相关的代谢产物离子。然而,我们开发的方法和OPLS-DA仅鉴定出4种离子,这表明基于代谢组学的数据分析方法设计的差异可能导致鉴定代谢物的差异。通过我们开发的方法和OPLS-DA共鉴定了20个与PIO结构相关的代谢物,其中6个是新的代谢物。结果表明,我们开发的两阶段数据分析方法可以有效地从相对复杂的基质中挖掘PIO代谢物离子的数据。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
6.30
自引率
2.80%
发文量
36
审稿时长
3.8 months
期刊介绍:
The journal aims to provide an international platform for scientists, researchers and academicians to promote, share and discuss new findings, current issues, and developments in the different areas of food and drug analysis.
The scope of the Journal includes analytical methodologies and biological activities in relation to food, drugs, cosmetics and traditional Chinese medicine, as well as related disciplines of topical interest to public health professionals.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


