De novo design of protein interactions with learned surface fingerprints
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 20
Abstract
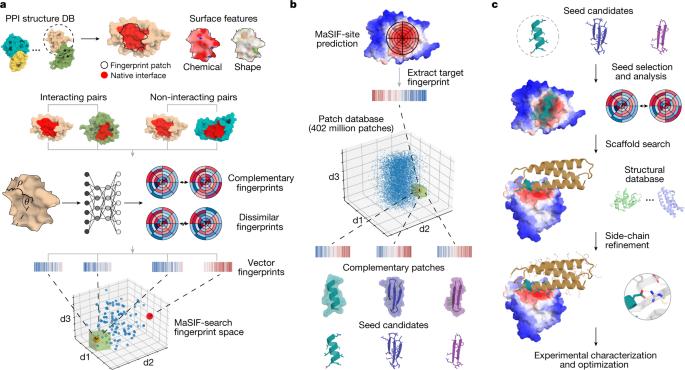
Physical interactions between proteins are essential for most biological processes governing life1. However, the molecular determinants of such interactions have been challenging to understand, even as genomic, proteomic and structural data increase. This knowledge gap has been a major obstacle for the comprehensive understanding of cellular protein–protein interaction networks and for the de novo design of protein binders that are crucial for synthetic biology and translational applications2–9. Here we use a geometric deep-learning framework operating on protein surfaces that generates fingerprints to describe geometric and chemical features that are critical to drive protein–protein interactions10. We hypothesized that these fingerprints capture the key aspects of molecular recognition that represent a new paradigm in the computational design of novel protein interactions. As a proof of principle, we computationally designed several de novo protein binders to engage four protein targets: SARS-CoV-2 spike, PD-1, PD-L1 and CTLA-4. Several designs were experimentally optimized, whereas others were generated purely in silico, reaching nanomolar affinity with structural and mutational characterization showing highly accurate predictions. Overall, our surface-centric approach captures the physical and chemical determinants of molecular recognition, enabling an approach for the de novo design of protein interactions and, more broadly, of artificial proteins with function. A surface-centric approach captures the physical and chemical determinants of molecular recognition, enabling the de novo design of protein interactions and of artificial proteins with function.
利用学习到的表面指纹从头设计蛋白质相互作用
蛋白质之间的物理相互作用对于支配生命的大多数生物过程都至关重要1。然而,即使基因组、蛋白质组和结构数据不断增加,要了解这种相互作用的分子决定因素却一直是个挑战。这一知识空白一直是全面了解细胞蛋白质-蛋白质相互作用网络的主要障碍,也是重新设计对合成生物学和转化应用至关重要的蛋白质结合体的主要障碍2-9。在这里,我们使用了一种在蛋白质表面运行的几何深度学习框架,它能生成指纹来描述对驱动蛋白质-蛋白质相互作用至关重要的几何和化学特征10。我们假设,这些指纹能捕捉分子识别的关键方面,代表了新型蛋白质相互作用计算设计的新范式。作为原理验证,我们通过计算设计了几种新的蛋白质结合剂,以与四种蛋白质靶标结合:SARS-CoV-2穗状病毒、PD-1、PD-L1和CTLA-4。有几种设计经过了实验优化,而另一些设计则是纯粹在硅学中生成的,亲和力达到了纳摩尔级,结构和突变特征显示了高度准确的预测。总之,我们以表面为中心的方法捕捉到了分子识别的物理和化学决定因素,为重新设计蛋白质相互作用以及更广泛意义上的具有功能的人工蛋白质提供了方法。以表面为中心的方法捕捉到了分子识别的物理和化学决定因素,从而能够重新设计蛋白质相互作用和具有功能的人工蛋白质。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


