Somatic genomic changes in single Alzheimer’s disease neurons
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 62
Abstract
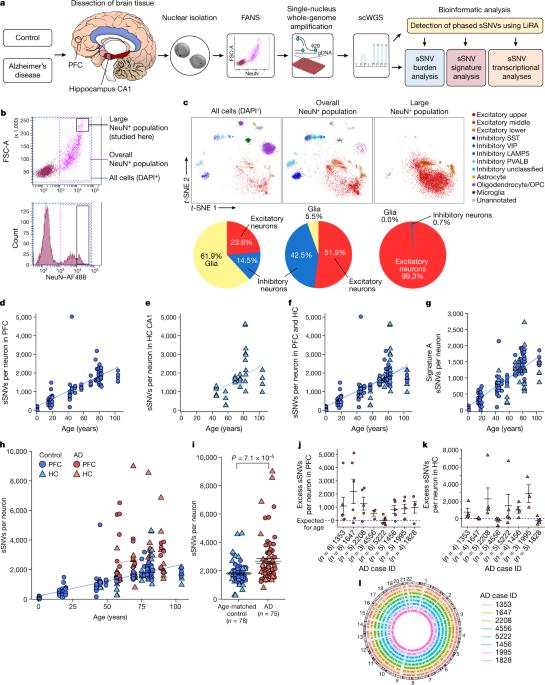
Dementia in Alzheimer’s disease progresses alongside neurodegeneration1–4, but the specific events that cause neuronal dysfunction and death remain poorly understood. During normal ageing, neurons progressively accumulate somatic mutations5 at rates similar to those of dividing cells6,7 which suggests that genetic factors, environmental exposures or disease states might influence this accumulation5. Here we analysed single-cell whole-genome sequencing data from 319 neurons from the prefrontal cortex and hippocampus of individuals with Alzheimer’s disease and neurotypical control individuals. We found that somatic DNA alterations increase in individuals with Alzheimer’s disease, with distinct molecular patterns. Normal neurons accumulate mutations primarily in an age-related pattern (signature A), which closely resembles ‘clock-like’ mutational signatures that have been previously described in healthy and cancerous cells6–10. In neurons affected by Alzheimer’s disease, additional DNA alterations are driven by distinct processes (signature C) that highlight C>A and other specific nucleotide changes. These changes potentially implicate nucleotide oxidation4,11, which we show is increased in Alzheimer’s-disease-affected neurons in situ. Expressed genes exhibit signature-specific damage, and mutations show a transcriptional strand bias, which suggests that transcription-coupled nucleotide excision repair has a role in the generation of mutations. The alterations in Alzheimer’s disease affect coding exons and are predicted to create dysfunctional genetic knockout cells and proteostatic stress. Our results suggest that known pathogenic mechanisms in Alzheimer’s disease may lead to genomic damage to neurons that can progressively impair function. The aberrant accumulation of DNA alterations in neurodegeneration provides insight into the cascade of molecular and cellular events that occurs in the development of Alzheimer’s disease. Analyses of single-cell whole-genome sequencing data show that somatic mutations are increased in the brain of individuals with Alzheimer’s disease compared to neurotypical individuals, with a pattern of genomic damage distinct from that of normal ageing.
单个阿尔茨海默病神经元的体细胞基因组变化。
阿尔茨海默病中的痴呆伴神经变性进展1-4,但导致神经元功能障碍和死亡的具体事件仍然知之甚少。在正常的衰老过程中,神经元以与分裂细胞相似的速度逐渐积累体细胞突变,这表明遗传因素、环境暴露或疾病状态可能影响这种积累。在这里,我们分析了来自阿尔茨海默病患者和神经正常对照个体的前额叶皮层和海马体的319个神经元的单细胞全基因组测序数据。我们发现阿尔茨海默病患者的体细胞DNA改变增加,具有独特的分子模式。正常神经元主要以与年龄相关的模式(特征A)积累突变,这与先前在健康细胞和癌细胞中描述的“时钟样”突变特征非常相似6-10。在受阿尔茨海默病影响的神经元中,额外的DNA改变是由不同的过程(特征C)驱动的,该过程突出了cbbbba和其他特定的核苷酸变化。这些变化可能与核苷酸氧化有关,我们发现,在阿尔茨海默病影响的神经元中,核苷酸氧化在原位增加。表达的基因表现出特征特异性损伤,突变表现出转录链偏倚,这表明转录偶联核苷酸切除修复在突变的产生中起作用。阿尔茨海默病的改变影响编码外显子,预计会产生功能失调的基因敲除细胞和蛋白质抑制应激。我们的研究结果表明,已知的阿尔茨海默病致病机制可能导致神经元的基因组损伤,从而逐渐损害其功能。神经退行性疾病中DNA改变的异常积累提供了对阿尔茨海默病发展过程中发生的分子和细胞级联事件的深入了解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


