A high-quality genome compendium of the human gut microbiome of Inner Mongolians
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 4
Abstract
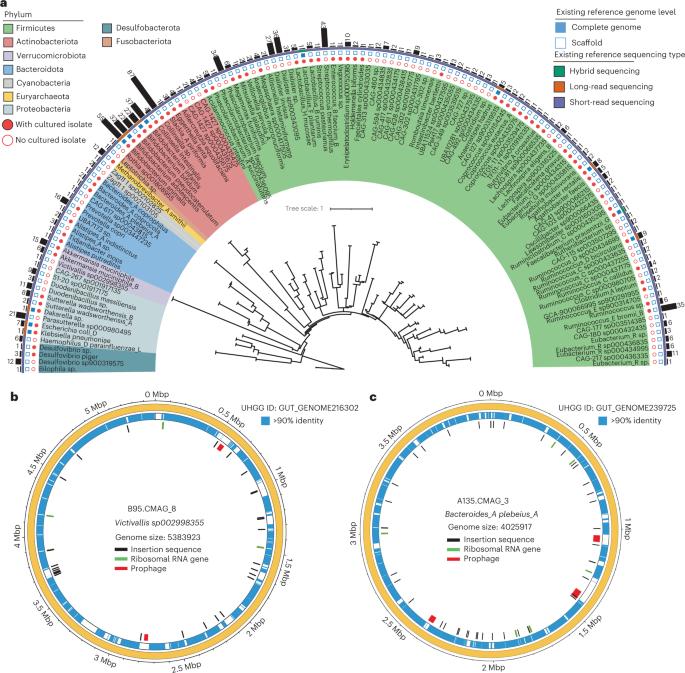
Metagenome-based resources have revealed the diversity and function of the human gut microbiome, but further understanding is limited by insufficient genome quality and a lack of samples from typically understudied populations. Here we used hybrid long-read PromethION and short-read HiSeq sequencing to characterize the faecal microbiota of 60 Inner Mongolian individuals (n = 180 samples over three time points) who were part of a probiotic yogurt intervention trial. We present the Inner Mongolian Gut Genome catalogue, comprising 802 closed and 5,927 high-quality metagenome-assembled genomes. This approach achieved high genome continuity and substantially increased the resolution of genomic elements, including ribosomal RNA operons, metabolic gene clusters, prophages and insertion sequences. Particularly, we report the ribosomal RNA operon copy numbers for uncultured species, over 12,000 previously undescribed gut prophages and the distribution of insertion sequence elements across gut bacteria. Overall, these data provide a high-quality, large-scale resource for studying the human gut microbiota. Hybrid long- and short-read metagenomic sequencing provides a high-resolution overview of genomic elements in the human gut microbiome.
高质量的内蒙古人肠道微生物组基因组汇编
基于元基因组的资源揭示了人类肠道微生物群的多样性和功能,但由于基因组质量不足和缺乏典型的未充分研究人群的样本,进一步的了解受到了限制。在这里,我们使用混合长读数 PromethION 和短读数 HiSeq 测序技术,对参加益生菌酸奶干预试验的 60 名内蒙古人(3 个时间点共 180 个样本)的粪便微生物群进行了表征。我们展示了内蒙古肠道基因组目录,其中包括 802 个封闭基因组和 5927 个高质量元基因组。这种方法实现了基因组的高度连续性,并大大提高了基因组元素的分辨率,包括核糖体 RNA 操作子、代谢基因簇、噬菌体和插入序列。特别是,我们报告了未培养物种的核糖体 RNA 操作子拷贝数、12,000 多个以前未描述过的肠道噬菌体以及插入序列元素在肠道细菌中的分布。总之,这些数据为研究人类肠道微生物群提供了高质量、大规模的资源。混合长短线程元基因组测序提供了人类肠道微生物群基因组元素的高分辨率概览。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


