A comprehensive map of molecular drug targets
IF 122.7
1区 医学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 1389
Abstract
The success of mechanism-based drug discovery depends on the definition of the drug target, but targets are often poorly defined in the literature. Here, Overington and colleagues present a comprehensive map of the molecular targets of approved drugs, and explore aspects including the footprint of target classes across disease areas, the success of privileged target families and drug target orthologues across standard model organisms. The success of mechanism-based drug discovery depends on the definition of the drug target. This definition becomes even more important as we try to link drug response to genetic variation, understand stratified clinical efficacy and safety, rationalize the differences between drugs in the same therapeutic class and predict drug utility in patient subgroups. However, drug targets are often poorly defined in the literature, both for launched drugs and for potential therapeutic agents in discovery and development. Here, we present an updated comprehensive map of molecular targets of approved drugs. We curate a total of 893 human and pathogen-derived biomolecules through which 1,578 US FDA-approved drugs act. These biomolecules include 667 human-genome-derived proteins targeted by drugs for human disease. Analysis of these drug targets indicates the continued dominance of privileged target families across disease areas, but also the growth of novel first-in-class mechanisms, particularly in oncology. We explore the relationships between bioactivity class and clinical success, as well as the presence of orthologues between human and animal models and between pathogen and human genomes. Through the collaboration of three independent teams, we highlight some of the ongoing challenges in accurately defining the targets of molecular therapeutics and present conventions for deconvoluting the complexities of molecular pharmacology and drug efficacy.
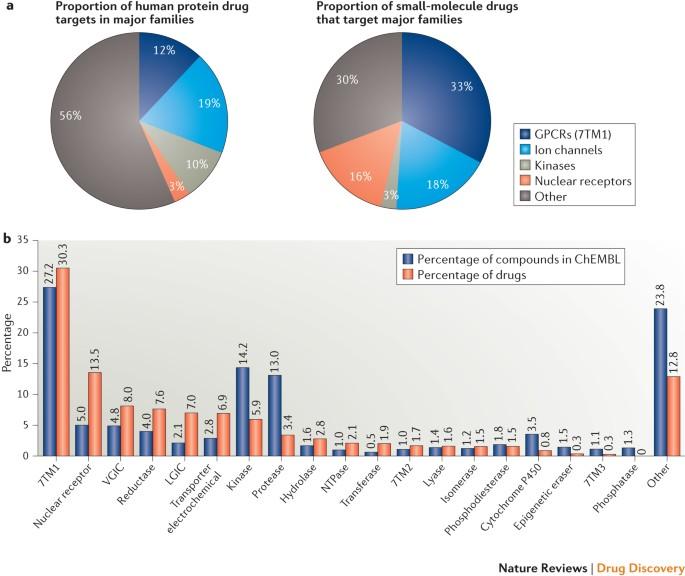
分子药物靶点综合地图
基于机理的药物发现的成功取决于药物靶点的定义,但文献中对靶点的定义往往很模糊。在这里,Overington及其同事展示了已获批准药物的分子靶点综合图谱,并探讨了包括靶点类别在不同疾病领域的分布、特权靶点家族的成功以及标准模式生物中的药物靶点同源物等方面的问题。基于机制的药物发现能否成功取决于药物靶点的定义。当我们试图将药物反应与遗传变异联系起来、了解分层临床疗效和安全性、合理解释同一类治疗药物之间的差异以及预测药物在患者亚群中的效用时,这一定义就变得更加重要。然而,文献中对药物靶点的定义往往不够明确,无论是已上市的药物还是正在研发的潜在治疗药物都是如此。在此,我们展示了已批准药物分子靶点的最新综合图谱。我们共整理了 893 种人类和病原体衍生的生物分子,1578 种美国 FDA 批准的药物通过这些生物分子发挥作用。这些生物分子包括 667 种人类疾病药物靶向的人类基因组衍生蛋白质。对这些药物靶点的分析表明,特权靶点家族在各个疾病领域继续占据主导地位,但新型的一流机制也在增长,尤其是在肿瘤学领域。我们探讨了生物活性类别与临床成功之间的关系,以及人类与动物模型之间、病原体与人类基因组之间是否存在同源物。通过三个独立团队的合作,我们强调了在准确定义分子疗法靶点方面正在面临的一些挑战,并提出了消除分子药理学和药物疗效复杂性的惯例。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Reviews. Drug Discovery
医学-生物工程与应用微生物
CiteScore
137.40
自引率
0.30%
发文量
227
期刊介绍:
Nature Reviews Drug Discovery is a monthly journal aimed at everyone working in the drug discovery and development arena.
Each issue includes:
Highest-quality reviews and perspectives covering a broad scope.
News stories investigating the hottest topics in drug discovery.
Timely summaries of key primary research papers.
Concise updates on the latest advances in areas such as new drug approvals, patent law, and emerging industry trends and strategies.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


