From leaves to labels: Building modular machine learning networks for rapid herbarium specimen analysis with LeafMachine2
Abstract
Premise
Quantitative plant traits play a crucial role in biological research. However, traditional methods for measuring plant morphology are time consuming and have limited scalability. We present LeafMachine2, a suite of modular machine learning and computer vision tools that can automatically extract a base set of leaf traits from digital plant data sets.
Methods
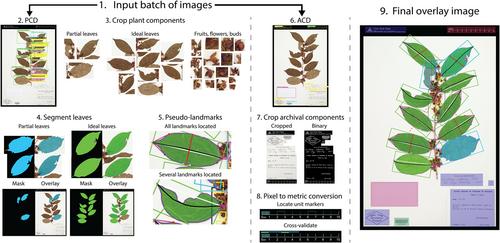
LeafMachine2 was trained on 494,766 manually prepared annotations from 5648 herbarium images obtained from 288 institutions and representing 2663 species; it employs a set of plant component detection and segmentation algorithms to isolate individual leaves, petioles, fruits, flowers, wood samples, buds, and roots. Our landmarking network automatically identifies and measures nine pseudo-landmarks that occur on most broadleaf taxa. Text labels and barcodes are automatically identified by an archival component detector and are prepared for optical character recognition methods or natural language processing algorithms.
Results
LeafMachine2 can extract trait data from at least 245 angiosperm families and calculate pixel-to-metric conversion factors for 26 commonly used ruler types.
Discussion
LeafMachine2 is a highly efficient tool for generating large quantities of plant trait data, even from occluded or overlapping leaves, field images, and non-archival data sets. Our project, along with similar initiatives, has made significant progress in removing the bottleneck in plant trait data acquisition from herbarium specimens and shifted the focus toward the crucial task of data revision and quality control.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


