Defining molecular basis for longevity traits in natural yeast isolates
IF 4.1
Q2 GERIATRICS & GERONTOLOGY
引用次数: 13
Abstract
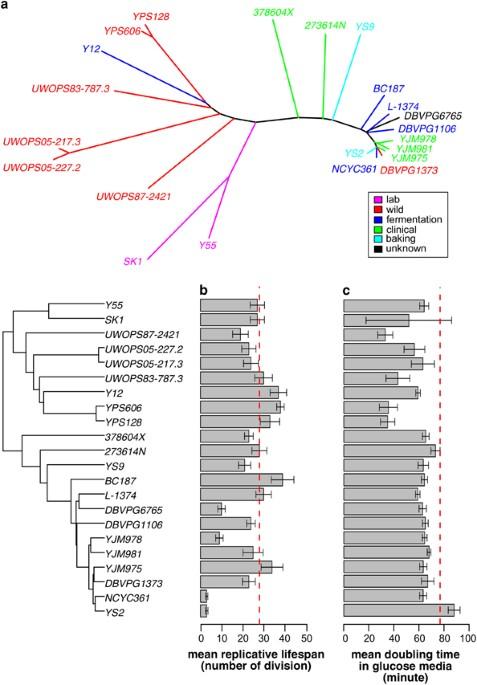
The budding yeast has served as a useful model organism in aging studies, leading to the identification of genetic determinants of longevity, many of which are conserved in higher eukaryotes. However, factors that promote longevity in a laboratory setting often have severe fitness disadvantages in the wild. To obtain an unbiased view on longevity regulation, we analyzed how a replicative lifespan is shaped by transcriptional, translational, metabolic, and morphological factors across 22 wild-type Saccharomyces cerevisiae isolates. We observed significant differences in lifespan across these strains and found that their longevity is strongly associated with up-regulation of oxidative phosphorylation and respiration and down-regulation of amino- acid and nitrogen compound biosynthesis. As calorie restriction and TOR signaling also extend the lifespan by adjusting many of the identified pathways, the data suggest that the natural plasticity of yeast lifespan is shaped by the processes that not only do not impose cost on fitness, but also are amenable to dietary intervention. A new study pinpoints a consistent set of genes and pathways underlying variations in yeast lifespan. Vadim Gladyshev at Harvard Medical School and co-workers analyzed 22 Saccharomyces cerevisiae yeast strains with diverse lifespans and habitats, looking to identify genomic signatures associated with natural variations in longevity. They observed a number of factors that characterized the longest-lived strains, including the upregulation of aerobic respiration, and found that interactions between genes and the environment were key. They also showed that factors associated with increased longevity in yeast strains do not necessarily degrade the fitness of those strains in the wild, and that longevity can be influenced through diet. The study thus paints a more complete picture of how environmental factors trigger changes—some hard–wired in the genome—that have real consequences on aging and longevity.
确定天然酵母分离物长寿特征的分子基础
在衰老研究中,芽殖酵母是一种有用的模式生物,它导致了长寿遗传决定因素的鉴定,其中许多因素在高等真核生物中是保守的。然而,在实验室环境中促进长寿的因素在野生环境中往往具有严重的适应性缺点。为了获得有关长寿调控的公正观点,我们分析了 22 个野生型酿酒酵母分离物的转录、翻译、代谢和形态因素是如何影响复制寿命的。我们观察到这些菌株的寿命存在显著差异,并发现它们的寿命与氧化磷酸化和呼吸的上调以及氨基酸和氮化合物生物合成的下调密切相关。由于卡路里限制和 TOR 信号转导也会通过调整许多已确定的通路来延长寿命,这些数据表明,酵母寿命的自然可塑性是由那些不仅不会对适应性造成代价,而且适合饮食干预的过程形成的。一项新研究确定了酵母寿命变化所依赖的一组一致的基因和途径。哈佛大学医学院的瓦迪姆-格拉迪舍夫(Vadim Gladyshev)及其合作者分析了22个具有不同寿命和栖息地的酿酒酵母菌株,希望找出与自然寿命变化相关的基因组特征。他们观察到最长寿菌株的一些特征因素,包括有氧呼吸的上调,并发现基因与环境之间的相互作用是关键。他们还发现,与酵母菌株寿命延长有关的因素并不一定会降低这些菌株在野生环境中的适应性,而且饮食也会影响它们的寿命。因此,这项研究更全面地描绘了环境因素如何引发变化--有些是基因组中的硬连接--从而对衰老和长寿产生真正的影响。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


