Systematic differences in discovery of genetic effects on gene expression and complex traits
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
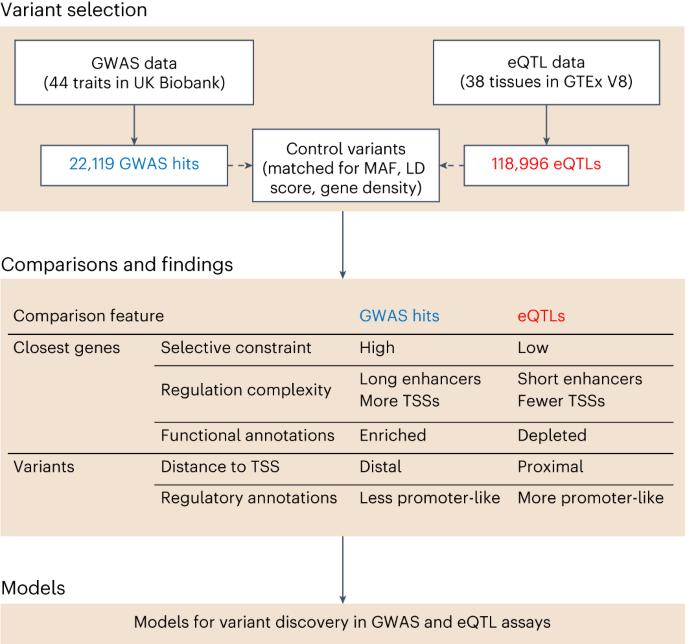
Most signals in genome-wide association studies (GWAS) of complex traits implicate noncoding genetic variants with putative gene regulatory effects. However, currently identified regulatory variants, notably expression quantitative trait loci (eQTLs), explain only a small fraction of GWAS signals. Here, we show that GWAS and cis-eQTL hits are systematically different: eQTLs cluster strongly near transcription start sites, whereas GWAS hits do not. Genes near GWAS hits are enriched in key functional annotations, are under strong selective constraint and have complex regulatory landscapes across different tissue/cell types, whereas genes near eQTLs are depleted of most functional annotations, show relaxed constraint, and have simpler regulatory landscapes. We describe a model to understand these observations, including how natural selection on complex traits hinders discovery of functionally relevant eQTLs. Our results imply that GWAS and eQTL studies are systematically biased toward different types of variant, and support the use of complementary functional approaches alongside the next generation of eQTL studies. This study seeks to explain the poor overlap of genome-wide association study and cis-expression quantitative trait locus variants using a model of differential selective constraint, suggesting that these two study types have biases towards different functional classes of variants.
发现基因表达和复杂性状的遗传效应的系统差异。
复杂性状的全基因组关联研究(GWAS)中的大多数信号都涉及具有假定基因调控作用的非编码遗传变异。然而,目前确定的调控变异,特别是表达数量性状基因座(eQTL),只能解释GWAS信号的一小部分。在这里,我们发现GWAS和顺式eQTL命中是系统性不同的:eQTL在转录起始位点附近强烈聚集,而GWAS命中则没有。GWAS命中附近的基因富含关键功能注释,受到强烈的选择性约束,在不同组织/细胞类型中具有复杂的调控景观,而eQTL附近的基因缺乏大多数功能注释,表现出宽松的约束,并且具有更简单的调控景观。我们描述了一个模型来理解这些观察结果,包括复杂性状的自然选择如何阻碍功能相关eQTL的发现。我们的研究结果表明,GWAS和eQTL的研究系统地偏向于不同类型的变体,并支持在下一代eQTL研究的同时使用互补功能方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


