Multiplex PCR-based molecular diagnostic method to detect cyantraniliprole-resistant I4790K mutation in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae)
Abstract
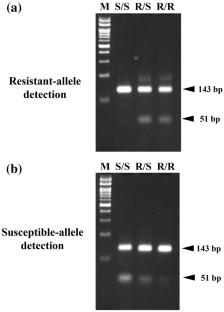
The diamondback moth Plutella xylostella (L.) (Lepidoptera: Plutellidae) is one of the most destructive insect pests worldwide which has developed resistance to many insecticides. Previously, a point mutation (I4790K) in a ryanodine receptor gene, a target gene of diamides, was identified as a major factor of resistance of the diamondback moth to cyantraniliprole in Japan. Although the diamondback moths have not yet widely developed resistance to cyantraniliprole in Japan, increasing resistant diamondback moth populations with the I4790K mutation in the future is a concern. To simply and quickly monitor the frequency of the I4790K mutations in field populations, we developed a multiplex polymerase chain reaction (PCR)-based molecular diagnostic method that can identify a genotype of the I4790K mutation site. We evaluated the sensitivity and specificity of the diagnostic method by comparing its cyantraniliprole-resistant and cyantraniliprole-susceptible allele detection results with those by Illumina MiSeq sequencing data. The results show sufficiently high sensitivity and specificity. Moreover, cyantraniliprole-resistant allele frequencies calculated by the molecular diagnostic method were almost comparable with those by MiSeq sequencing data. The molecular diagnostic method would help in performing continuous monitoring of the cyantraniliprole resistance level of diamondback moth populations in the field.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


