Functional activity of E. coli RNase R in the Antarctic Pseudomonas syringae Lz4W
IF 3.5
Q3 Biochemistry, Genetics and Molecular Biology
Journal of Genetic Engineering and Biotechnology
Pub Date : 2023-12-01
DOI:10.1186/s43141-023-00553-2
引用次数: 0
Abstract
Background
In Antarctic P. syringae RNase R play an essential role in the processing of 16S and 5S rRNA, thereby playing an important role in cold-adapted growth of the bacterium. This study is focused on deciphering the in vivo functional activity of mesophilic exoribonuclease R and its catalytic domain (RNB) in an evolutionary distant psychrophilic bacterium Pseudomonas syringae Lz4W.
Results
Our results confirm that E. coli RNase R complemented the physiological functions of the psychrophilic bacterium P. syringae RNase R and rescued the cold-sensitive phenotype of Pseudomonas syringae ∆rnr mutant. More importantly, the catalytic domain (RNB) of the E. coli RNase R is also capable of alleviating the cold-sensitive growth defects of ∆rnr mutant as seen with the catalytic domain (RNB) of the P. syringae enzyme. The Catalytic domain of E. coli RNase R was less efficient than the Catalytic domain of P. syringae RNase R in rescuing the cold-sensitive growth of ∆rnr mutant at 4°C, as the ∆rnr expressing the RNBEc (catalytic domain of E. coli RNase R) displayed longer lag phase than the RNBPs (Catalytic domain of P. syringae RNase R) complemented ∆rnr mutant at 4°C. Altogether it appears that the E. coli RNase R and P. syringae RNase R are functionally exchangeable for the growth requirements of P. syringae at low temperature (4°C). Our results also confirm that in P. syringae the requirement of RNase R for supporting the growth at 4°C is independent of the degradosomal complex.
Conclusion
E. coli RNase R (RNase REc) rescues the cold-sensitive phenotype of the P. syringae Δrnr mutant. Similarly, the catalytic domain of E. coli RNase R (RNBEc) is also capable of supporting the growth of Δrnr mutant at low temperatures. These findings have a vast scope in the design and development of low-temperature-based expression systems.
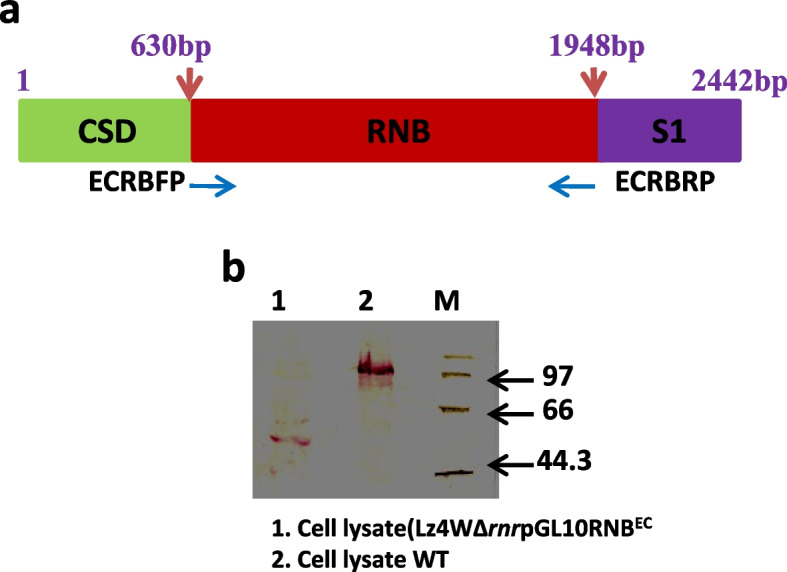
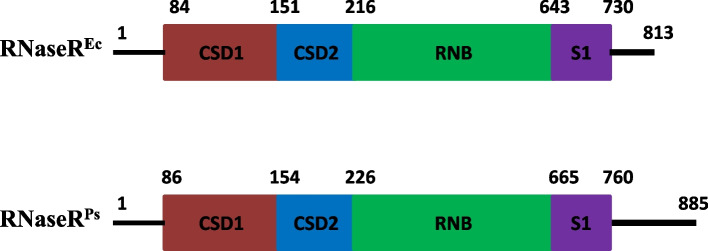
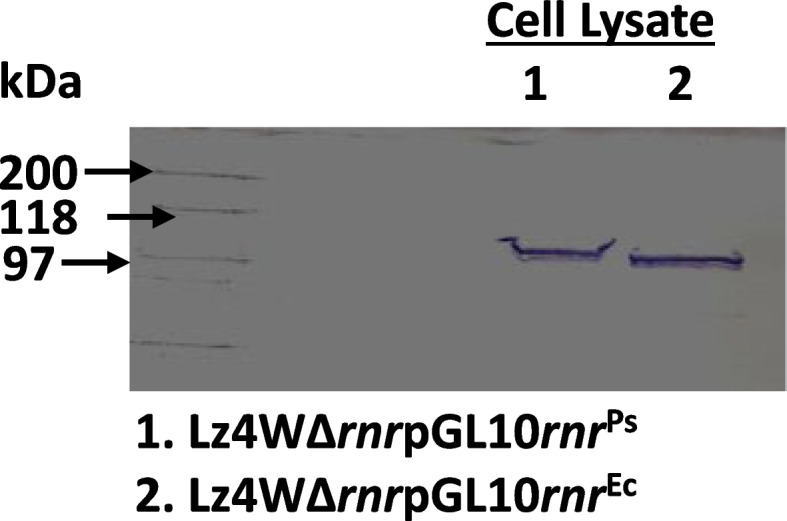
南极丁香假单胞菌Lz4W中大肠杆菌RNase R的功能活性。
背景:在南极,丁香RNase R在16S和5S rRNA的加工中起着重要作用,从而在细菌的冷适应生长中发挥着重要作用。本研究的重点是在一种进化遥远的嗜冷细菌Pseudomonas syringae Lz4W中解读嗜中温外核糖核酸酶R及其催化结构域(RNB)的体内功能活性丁香假单胞菌∆rnr突变体。更重要的是,大肠杆菌核糖核酸酶R的催化结构域(RNB)也能够减轻∆rnr突变体的冷敏感生长缺陷,正如丁香花假单胞菌酶的催化结构区(RNB)所见。在4°C下,大肠杆菌RNase R的催化结构域在拯救∆rnr突变体的冷敏生长方面不如丁香花RNase R催化结构域有效,因为表达RNBEc(大肠杆菌RNaseR催化结构区)的∆rnr在4°C.时比RNBPs(丁香花RNaseR的催化域)补充的∆rnr突变体显示出更长的滞后期。总之,在低温(4°C)下,大肠杆菌RNase R和丁香花RNase R在功能上可交换丁香花的生长需求。我们的结果还证实,在丁香中,支持4°C生长的RNase R的需求与脱颗粒体复合体无关。结论:大肠杆菌RNase R(RNase REc)挽救了丁香花Δrnr突变体的冷敏表型。同样,大肠杆菌核糖核酸酶R(RNBEc)的催化结构域也能够在低温下支持Δrnr突变体的生长。这些发现在基于低温的表达系统的设计和开发中具有广阔的范围。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Genetic Engineering and Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
5.70
自引率
5.70%
发文量
159
审稿时长
16 weeks
期刊介绍:
Journal of genetic engineering and biotechnology is devoted to rapid publication of full-length research papers that leads to significant contribution in advancing knowledge in genetic engineering and biotechnology and provide novel perspectives in this research area. JGEB includes all major themes related to genetic engineering and recombinant DNA. The area of interest of JGEB includes but not restricted to: •Plant genetics •Animal genetics •Bacterial enzymes •Agricultural Biotechnology, •Biochemistry, •Biophysics, •Bioinformatics, •Environmental Biotechnology, •Industrial Biotechnology, •Microbial biotechnology, •Medical Biotechnology, •Bioenergy, Biosafety, •Biosecurity, •Bioethics, •GMOS, •Genomic, •Proteomic JGEB accepts
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


