Population stratification correction using Bayesian shrinkage priors for genetic association studies
Abstract
Introduction
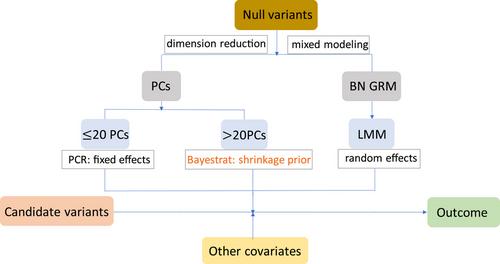
Population stratification (PS) is a major source of confounding in population-based genetic association studies of quantitative traits. Principal component regression (PCR) and linear mixed model (LMM) are two commonly used approaches to account for PS in association studies. Previous studies have shown that LMM can be interpreted as including all principal components (PCs) as random-effect covariates. However, including all PCs in LMM may dilute the influence of relevant PCs in some scenarios, while including only a few preselected PCs in PCR may fail to fully capture the genetic diversity.
Materials and methods
To address these shortcomings, we introduce Bayestrat—a method to detect associated variants with PS correction under the Bayesian LASSO framework. To adjust for PS, Bayestrat accommodates a large number of PCs and utilizes appropriate shrinkage priors to shrink the effects of nonassociated PCs.
Results
Simulation results show that Bayestrat consistently controls type I error rates and achieves higher power compared to its non-shrinkage counterparts, especially when the number of PCs included in the model is large. As a demonstration of the utility of Bayestrat, we apply it to the Multi-Ethnic Study of Atherosclerosis (MESA). Variants and genes associated with serum triglyceride or HDL cholesterol are identified in our analyses.
Discussion
The automatic and self-selection features of Bayestrat make it particularly suited in situations with complex underlying PS scenarios, where it is unknown a priori which PCs are potential confounders, yet the number that needs to be considered could be large in order to fully account for PS.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


