Preparing ductal epithelial organoids for high-spatial-resolution molecular profiling using mass spectrometry imaging
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 6
Abstract
Organoid culture systems are self-renewing, three-dimensional (3D) models derived from pluripotent stem cells, adult derived stem cells or cancer cells that recapitulate key molecular and structural characteristics of their tissue of origin. They generally form into hollow structures with apical–basolateral polarization. Mass spectrometry imaging (MSI) is a powerful analytical method for detecting a wide variety of molecules in a single experiment while retaining their spatiotemporal distribution. Here we describe a protocol for preparing organoids for MSI that (1) preserves the 3D morphological structure of hollow organoids, (2) retains the spatiotemporal distribution of a vast array of molecules (3) and enables accurate molecular identification based on tandem mass spectrometry. The protocol specifically focuses on the collection and embedding of the organoids in gelatin, and gives recommendations for MSI-specific sample preparation, data acquisition and molecular identification by tandem mass spectrometry. This method is applicable to a wide range of organoids from different origins, and takes 1 d from organoid collection to MSI data acquisition. Organoids are increasingly used as model systems in precision medicine, but they are delicate, making it difficult to get accurate spatial chemical information. This protocol describes how to prepare organoid samples for mass spectrometry imaging.
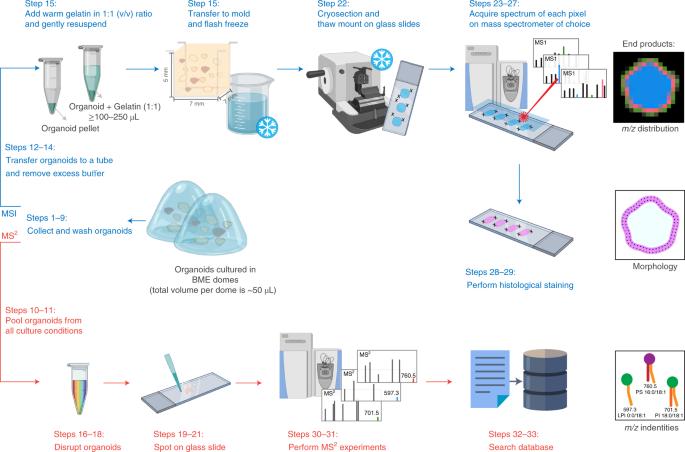
利用质谱成像技术制备用于高空间分辨率分子图谱分析的导管上皮细胞器官组织
类器官培养系统是一种可自我更新的三维(3D)模型,由多能干细胞、成体干细胞或癌细胞衍生而来,可再现其原生组织的关键分子和结构特征。它们通常形成具有顶端-基底极化的中空结构。质谱成像(MSI)是一种功能强大的分析方法,可在一次实验中检测多种分子,同时保留其时空分布。在此,我们介绍了一种制备用于 MSI 的有机体的方案,该方案(1)保留了空心有机体的三维形态结构,(2)保留了大量分子的时空分布(3)并能基于串联质谱进行准确的分子鉴定。该方案特别关注明胶中有机体的收集和包埋,并对 MSI 特定样品的制备、数据采集和串联质谱的分子鉴定提出了建议。该方法适用于各种不同来源的有机体,从收集有机体到获取 MSI 数据只需 1 天。有机体越来越多地被用作精准医学的模型系统,但它们很脆弱,很难获得准确的空间化学信息。本规程介绍了如何为质谱成像制备类器官样本。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


