Automatic parcellation of resting-state cortical dynamics by iterative community detection and similarity measurements.
IF 3.1
Q2 NEUROSCIENCES
AIMS Neuroscience
Pub Date : 2021-09-10
eCollection Date: 2021-01-01
DOI:10.3934/Neuroscience.2021028
引用次数: 2
Abstract
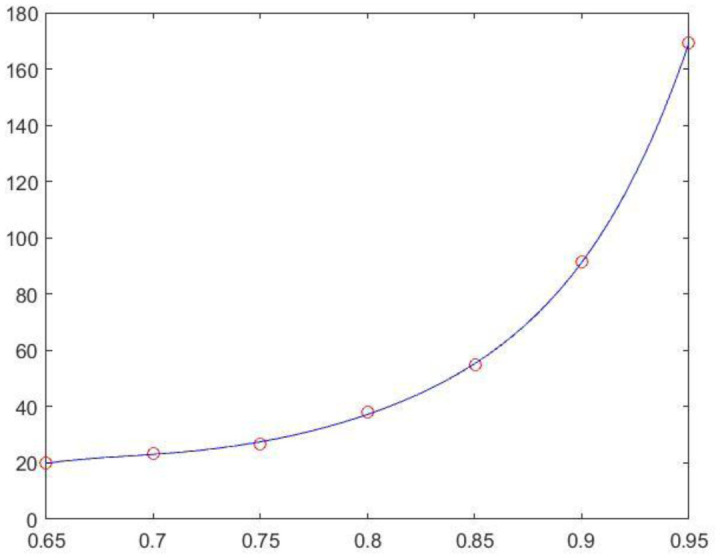
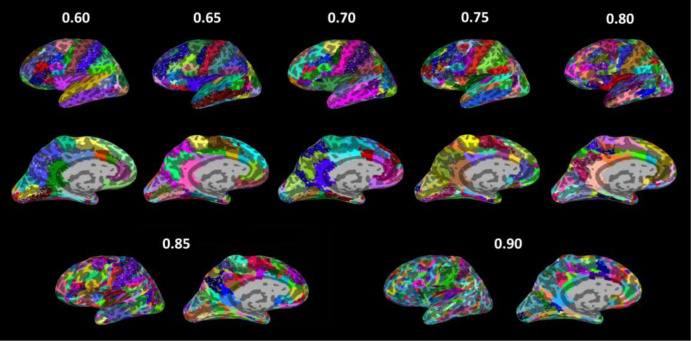
To investigate the properties of a large-scale brain network, it is a common practice to reduce the dimension of resting state functional magnetic resonance imaging (rs-fMRI) data to tens to hundreds of nodes. This study presents an analytic streamline that incorporates modular analysis and similarity measurements (MOSI) to fulfill functional parcellation (FP) of the cortex. MOSI is carried out by iteratively dividing a module into sub-modules (via the Louvain community detection method) and unifying similar neighboring sub-modules into a new module (adjacent sub-modules with a similarity index <0.05) until the brain modular structures of successive runs become constant. By adjusting the gamma value, a parameter in the Louvain algorithm, MOSI may segment the cortex with different resolutions. rs-fMRI scans of 33 healthy subjects were selected from the dataset of the Rockland sample. MOSI was applied to the rs-fMRI data after standardized pre-processing steps. The results indicate that the parcellated modules by MOSI are more homogeneous in content. After reducing the grouped voxels to representative neural nodes, the network structures were explored. The resultant network components were comparable with previous reports. The validity of MOSI in achieving data reduction has been confirmed. MOSI may provide a novel starting point for further investigation of the network properties of rs-fMRI data. Potential applications of MOSI are discussed.
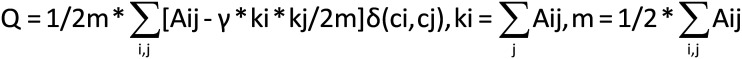
基于迭代群落检测和相似性测量的静息状态皮质动态自动分割。
为了研究大规模脑网络的特性,将静息状态功能磁共振成像(rs-fMRI)数据降维到几十到几百个节点是一种常见的做法。本研究提出了一种分析流线,结合模块化分析和相似性测量(MOSI)来实现皮质的功能分割(FP)。MOSI是通过将一个模块迭代划分为子模块(通过Louvain社区检测方法),并将相似的相邻子模块统一为一个新的模块(具有相似指数的相邻子模块)来实现的
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

AIMS Neuroscience
NEUROSCIENCES-
CiteScore
4.20
自引率
0.00%
发文量
26
审稿时长
8 weeks
期刊介绍:
AIMS Neuroscience is an international Open Access journal devoted to publishing peer-reviewed, high quality, original papers from all areas in the field of neuroscience. The primary focus is to provide a forum in which to expedite the speed with which theoretical neuroscience progresses toward generating testable hypotheses. In the presence of current and developing technology that offers unprecedented access to functions of the nervous system at all levels, the journal is designed to serve the role of providing the widest variety of the best theoretical views leading to suggested studies. Single blind peer review is provided for all articles and commentaries.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


