The identification of novel promoters and terminators for protein expression and metabolic engineering applications in Kluyveromyces marxianus
Abstract
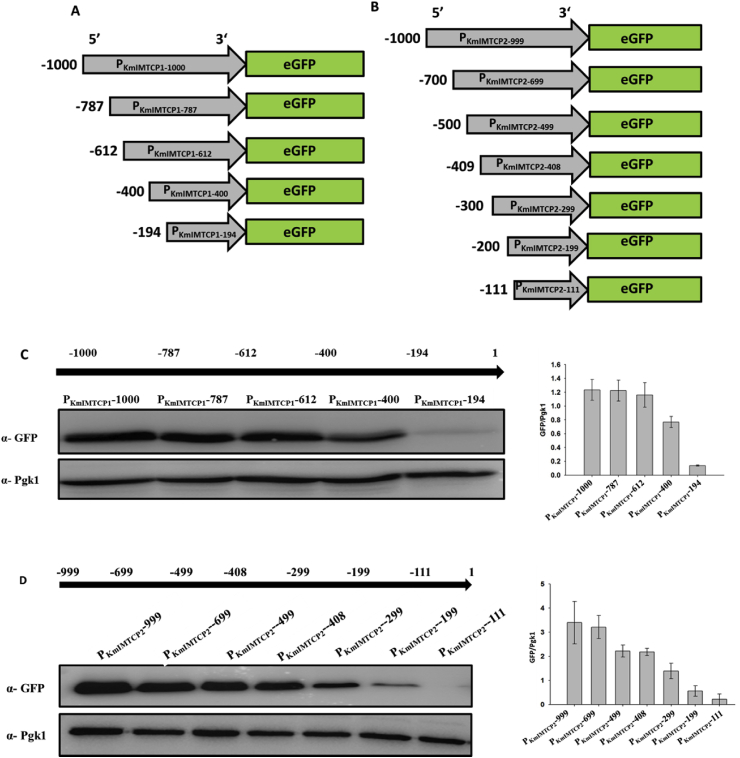
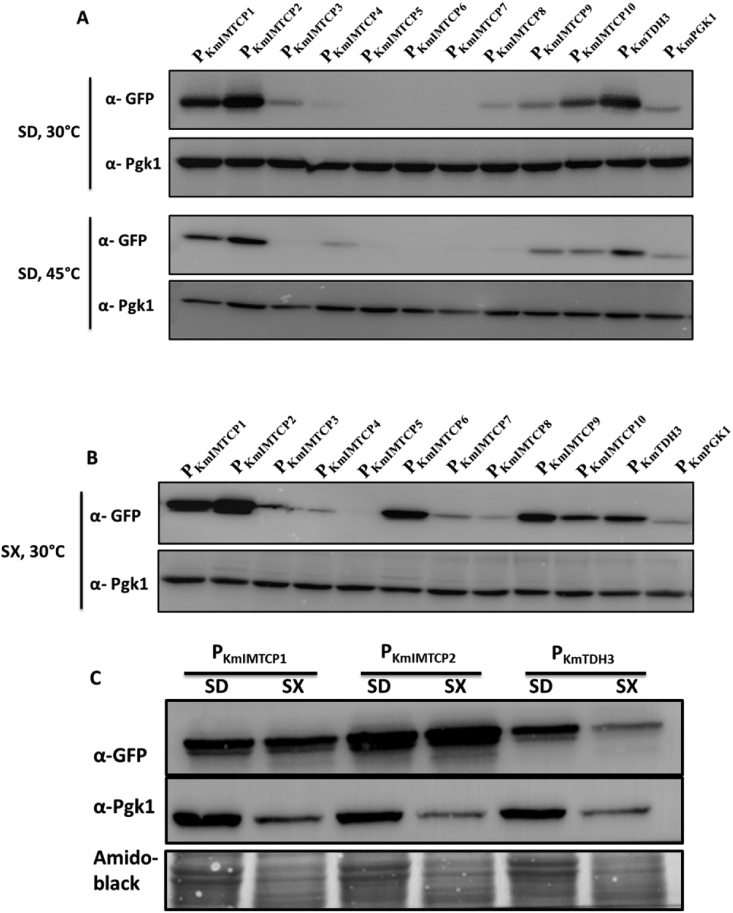
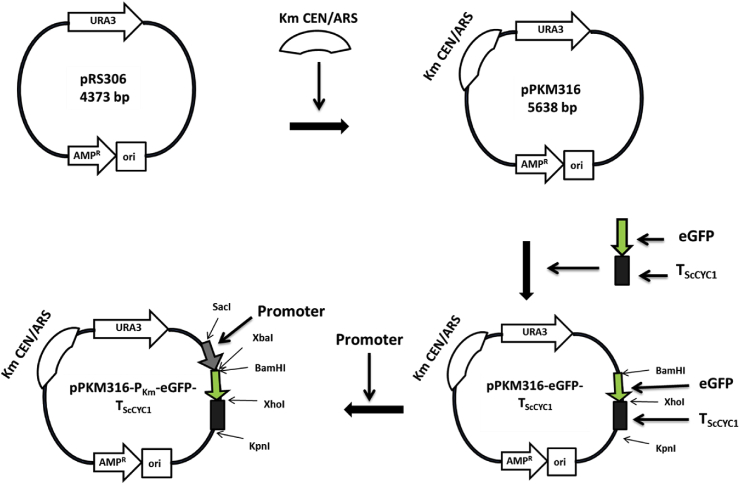
The K. marxianus has emerged as a potential yeast strain for various biotechnological applications. However, the limited number of available genetic tools has hindered the widespread usage of this yeast. In the current study we have expanded the molecular tool box by identifying novel sets of promoters and terminators for increased recombinant protein expression in K. marxianus. The previously available transcriptomic data were analyzed to identify top 10 promoters of highest gene expression activity. We further characterized and compared strength of these identified promoters using eGFP as a reporter protein, at different temperatures and carbon sources. To examine the regulatory region driving protein expression, serially truncated shorter versions of two selected strong promoters were designed, and examined for their ability to drive eGFP protein expression. The activities of these two promoters were further enhanced using different combinations of native transcription terminators of K. marxianus. We further utilized the identified DNA cassette encoding strong promoter in metabolic engineering of K. marxianus for enhanced β-galactosidase activity. The present study thus provides novel sets of promoters and terminators as well as engineered K. marxianus strain for its wider utility in applications requiring lactose degradation such as in cheese whey and milk.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


