Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 102
Abstract
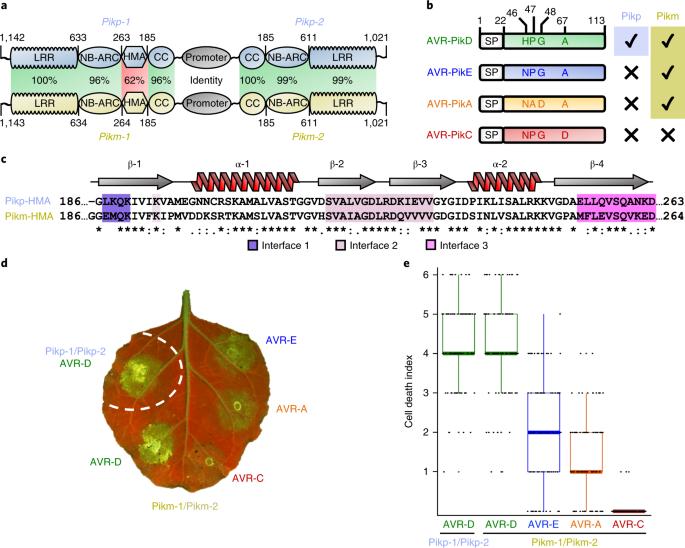
Accelerated adaptive evolution is a hallmark of plant–pathogen interactions. Plant intracellular immune receptors (NLRs) often occur as allelic series with differential pathogen specificities. The determinants of this specificity remain largely unknown. Here, we unravelled the biophysical and structural basis of expanded specificity in the allelic rice NLR Pik, which responds to the effector AVR-Pik from the rice blast pathogen Magnaporthe oryzae. Rice plants expressing the Pikm allele resist infection by blast strains expressing any of three AVR-Pik effector variants, whereas those expressing Pikp only respond to one. Unlike Pikp, the integrated heavy metal-associated (HMA) domain of Pikm binds with high affinity to each of the three recognized effector variants, and variation at binding interfaces between effectors and Pikp-HMA or Pikm-HMA domains encodes specificity. By understanding how co-evolution has shaped the response profile of an allelic NLR, we highlight how natural selection drove the emergence of new receptor specificities. This work has implications for the engineering of NLRs with improved utility in agriculture. NLR immune receptors recognize pathogen effectors and activate a response that leads to resistance. The specific interactions between five rice receptor variants and their cognate effectors are studied by solving the structures of the complexes.
水稻 NLR 中的多态残基扩大了与稻瘟病病原体效应因子的结合和反应
加速适应性进化是植物与病原体相互作用的一个标志。植物细胞内免疫受体(NLRs)往往是具有不同病原体特异性的等位基因系列。这种特异性的决定因素在很大程度上仍然未知。在这里,我们揭示了等位基因水稻 NLR Pik 扩大特异性的生物物理和结构基础,它对稻瘟病病原体 Magnaporthe oryzae 的效应物 AVR-Pik 有反应。表达 Pikm 等位基因的水稻植株能抵抗表达三种 AVR-Pik 效应体变体中任何一种的稻瘟病菌株的感染,而表达 Pikp 的植株只对其中一种有反应。与 Pikp 不同的是,Pikm 的集成重金属相关(HMA)结构域能与三种公认的效应物变体中的每一种高亲和力结合,效应物与 Pikp-HMA 或 Pikm-HMA 结构域之间结合界面的变异编码了特异性。通过了解共同进化如何塑造了等位基因 NLR 的反应特征,我们强调了自然选择如何推动了新受体特异性的出现。这项工作对改进农业实用性的 NLR 工程具有重要意义。NLR 免疫受体能识别病原体效应物,并激活导致抗性的反应。通过解析复合物的结构,研究了五种水稻受体变体与其同源效应物之间的特异性相互作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


