Omicron Subvariants, Including BA.4 and BA.5, Substantially Preserve T Cell Epitopes of Ancestral SARS-CoV-2.
IF 4.3
4区 医学
Q2 IMMUNOLOGY
引用次数: 6
Abstract
https://immunenetwork.org The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variants (B.1.1.529 and related) that emerged in November 2021 have spread worldwide, and are designated a variant of concern by the World Health Organization (WHO) (1). They have become the dominant strains, comprising >99% of newly deposited SARS-CoV-2 sequences in GISIAD (www. gisaid.org) as of May 16, 2022 (2). Following the emergence of B.1.1.529 (BA.1), BA.2 (often called stealth Omicron) soon became the most prevalent sublineage worldwide. Subsequent subvariants, including BA.2.9, BA.2.12.1, BA.4, and BA.5, are now rapidly dominating the circulating Omicron subvariants in originating regions, and are beginning to spread globally (2). The dominance of Omicron variants and their rapid evolution into various subvariants have raised concerns regarding the effects of the immunity elicited by natural infection or vaccination. As evolution continues, the variants’ spike proteins exhibit higher affinities toward ACE2, and/ or increasing capacity for evading preformed neutralizing antibodies induced by previous natural infection or vaccination (3-5). These changes are consistent with increased breakthrough infections and re-infections with Omicron variants (6,7).
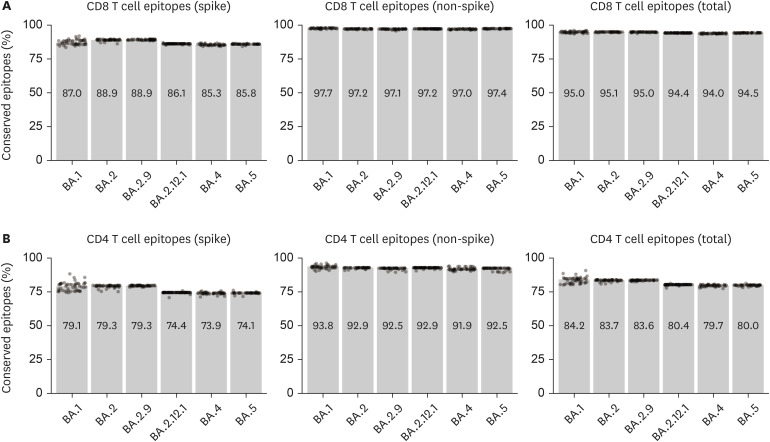
包括BA.4和BA.5在内的组粒亚变体大量保留了祖先SARS-CoV-2的T细胞表位。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Immune Network
Immunology and Microbiology-Immunology
CiteScore
2.90
自引率
3.30%
发文量
36
期刊介绍:
Immune Network publishes novel findings in basic and clinical immunology and aims to provide a medium through which researchers in various fields of immunology can share and connect. The journal focuses on advances and insights into the regulation of the immune system and the immunological mechanisms of various diseases. Research that provides integrated insights into translational immunology is given preference for publication. All submissions are evaluated based on originality, quality, clarity, and brevity
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


