Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 1109
Abstract
Jiayang Li and colleagues report the positional cloning of the Ideal Plant Architecture (IPA1) QTL in rice. The gene OsSPL14 underlies the IPA1 locus and regulates plant architecture and enhances rice grain yield. Increasing crop yield is a major challenge for modern agriculture. The development of new plant types, which is known as ideal plant architecture (IPA), has been proposed as a means to enhance rice yield potential over that of existing high-yield varieties1,2. Here, we report the cloning and characterization of a semidominant quantitative trait locus, IPA1 (Ideal Plant Architecture 1), which profoundly changes rice plant architecture and substantially enhances rice grain yield. The IPA1 quantitative trait locus encodes OsSPL14 (SOUAMOSA PROMOTER BINDING PROTEIN-LIKE 14) and is regulated by microRNA (miRNA) OsmiR156 in vivo. We demonstrate that a point mutation in OsSPL14 perturbs OsmiR156-directed regulation of OsSPL14, generating an ''ideal'' rice plant with a reduced tiller number, increased lodging resistance and enhanced grain yield. Our study suggests that OsSPL14 may help improve rice grain yield by facilitating the breeding of new elite rice varieties.
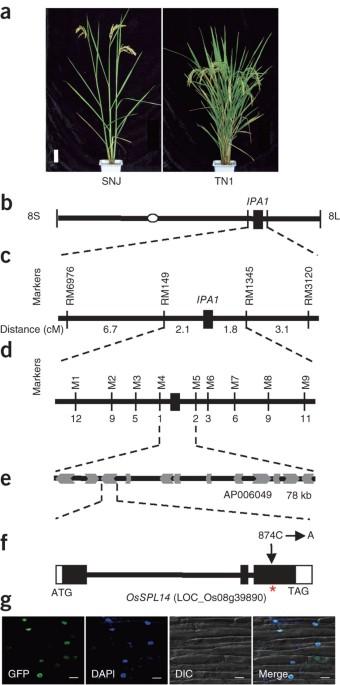
OsmiR156 对 OsSPL14 的调控决定了水稻的理想植株结构
李家洋及其同事报告了水稻理想植株结构(IPA1)QTL的定位克隆。基因 OsSPL14 是 IPA1 基因座的基础,它调控植物结构并提高水稻的产量。提高作物产量是现代农业面临的一大挑战。开发新的植株类型(即理想植株结构(IPA))被认为是提高现有高产品种水稻产量潜力的一种手段1,2。在此,我们报告了一个半显性数量性状基因座 IPA1(理想植株结构 1)的克隆和表征,该基因座可显著改变水稻植株结构,并大幅提高水稻籽粒产量。IPA1 数量性状基因座编码 OsSPL14(SOUAMOSA PROMOTER BINDING PROTEIN-LIKE 14),在体内受 microRNA(miRNA)OsmiR156 的调控。我们证明,OsSPL14 的点突变扰乱了 OsmiR156 对 OsSPL14 的定向调控,产生了一种 "理想 "水稻植株,其分蘖数减少,抗倒伏性增强,谷物产量提高。我们的研究表明,OsSPL14可能有助于培育新的水稻优良品种,从而提高水稻的产量。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


