Curvature-Sensing Peptides for Virus and Extracellular Vesicle Applications
IF 16
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
Membrane curvature is a key biophysical feature that regulates diverse biological processes. A wide range of natural proteins have distinct structural motifs that can sense membrane curvature and function independently as curvature-sensing peptides. In recent years, such peptides have demonstrated excellent potential for selectively capturing and disrupting enveloped viruses and extracellular vesicles (EVs), which are in the ∼30–300 nm size range and possess highly curved membranes. Despite extensive progress, there is an outstanding need to categorize different curvature-sensing motifs and link them to tailored peptide designs for specific applications. Herein, we introduce membrane curvature sensing as a unifying selectivity principle to target enveloped viruses and EVs, and critically evaluate the structural and mechanistic features of curvature-sensing motifs to develop a four-type classification framework that spans membrane binding and disruption. These efforts are supported by foundational studies and the latest insights obtained from experimental, theoretical, and simulation approaches as well as machine learning-aided thermodynamic modeling. According to this framework, we analyze recent advances to engineer curvature-sensing peptides for emerging applications such as one-step diagnostic capture, rapid virus quantification, antiviral and anti-EV therapies, and genome-wide screening of bacterial EV regulators. In turn, we illustrate how design parameters such as amino acid composition, conformational dynamics, and interfacial force balancing (e.g., electrostatics vs hydrophobicity) guide functional performance. These insights lead us to propose future directions for curvature-sensing peptide engineering and highlight outstanding scientific questions and translational opportunities.
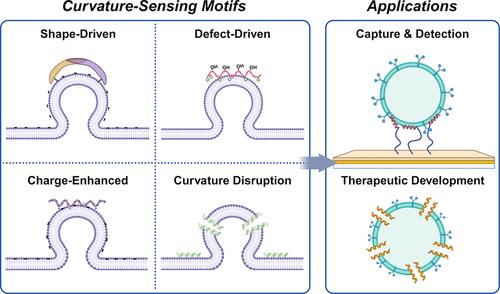
曲率传感肽在病毒和细胞外囊泡中的应用
膜曲率是调节多种生物过程的关键生物物理特征。广泛的天然蛋白质具有独特的结构基序,可以感知膜曲率并作为曲率传感肽独立发挥作用。近年来,这类肽在选择性捕获和破坏包膜病毒和细胞外囊泡(ev)方面表现出了极好的潜力,这些病毒和细胞外囊泡的尺寸在~ 30-300 nm范围内,具有高度弯曲的膜。尽管取得了广泛的进展,但仍然需要对不同的曲率传感基序进行分类,并将它们与特定应用的定制肽设计联系起来。在此,我们将膜曲率感知作为针对包膜病毒和ev的统一选择性原则,并批判性地评估了曲率感知基元的结构和机制特征,以建立跨越膜结合和破坏的四类分类框架。这些努力得到了基础研究和从实验、理论和模拟方法以及机器学习辅助热力学建模中获得的最新见解的支持。根据这一框架,我们分析了曲率传感肽工程的最新进展,用于诸如一步诊断捕获、快速病毒定量、抗病毒和抗EV治疗以及细菌EV调节因子的全基因组筛选等新兴应用。反过来,我们说明了设计参数如氨基酸组成、构象动力学和界面力平衡(例如,静电与疏水性)如何指导功能性能。这些见解引导我们提出曲率传感肽工程的未来方向,并突出突出的科学问题和转化机会。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


