Genomic insights into persistent infections, reinfections, and subspecies diversity of Mycobacteroides abscessus: A whole-genome sequencing study of Thai and global isolates
IF 2.6
4区 医学
Q3 INFECTIOUS DISEASES
引用次数: 0
Abstract
Mycobacteroides abscessus is a highly resistant pathogen with significant genetic diversity and complicating clinical management. This study used whole-genome sequencing to analyze genomic variations in seven serially collected isolates from three Thai patients, distinguishing between persistent infections and reinfections. Comparative analysis with 43 global isolates revealed subspecies-specific genetic diversity and distribution patterns. Among Thai isolates, two paired samples (P7.1-P7.2 and P15.1-P15.2) were persistent infections (same clone), while two (P8.1-P8.2 and P15.2-P15.3) were reinfections (different clones). Genome-wide comparisons revealed depth distribution patterns and gene cluster variations among different clones, whereas minimal divergence was observed within persistent infections. Although initial pan-genome analysis identified unique genes in same-clone pairs (P7.1 vs. P7.2 and P15.1 vs. P15.2), further validation using raw read mapping confirmed these genes were not truly unique. Analysis of global isolates showed subspecies-specific genetic variations. M. abscessus subsp. abscessus (MAB) and M. abscessus subsp. massiliense (MMAS) exhibited distinct genotypically drug resistance profiles, with unique core genes linked to adaptation and resistance mechanisms. STRING analysis identified 42 unique core genes in MAB, with 11 gene interactions—truB, prmC_2, and aguA_2 showing the highest interaction scores. In contrast, MMAS had 11 unique core genes with a single interaction between lgrD_4 and rnc. Subsequent validation using NCBI BLAST showed only fmt_2 and aguA_2 were truly unique to MAB. This study provides new insights into the genomic evolution of M. abscessus during persistent and reinfections and genetic variation among subspecies. The findings enhance understanding of M. abscessus epidemiology and may inform therapeutic and infection control.
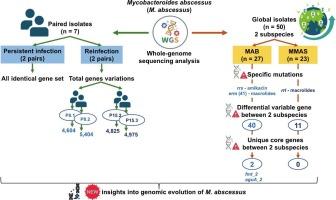
持续感染、再感染和脓肿分枝杆菌亚种多样性的基因组研究:泰国和全球分离株的全基因组测序研究。
脓肿分枝杆菌是一种高度耐药的病原体,具有显著的遗传多样性和复杂的临床管理。该研究使用全基因组测序分析了从3名泰国患者连续收集的7株分离株的基因组变异,区分了持续感染和再感染。与43个全球分离株的比较分析揭示了亚种特异性遗传多样性和分布格局。在泰国分离株中,2对样本(p7.1 ~ p7.2和p15.1 ~ p15.2)为持续感染(同一克隆),2对样本(p8.1 ~ p8.2和p15.2 ~ p15.3)为再感染(不同克隆)。全基因组比较揭示了不同克隆之间的深度分布模式和基因簇变异,而在持续感染中观察到的差异很小。虽然最初的泛基因组分析确定了相同克隆对中的独特基因(P7.1 vs. P7.2和P15.1 vs. P15.2),但使用原始读取图谱进一步验证证实这些基因并非真正独特。全球分离株分析显示亚种特异性遗传变异。脓肿支原体脓疡芽孢杆菌(MAB)和脓疡芽孢杆菌。马尾蛇(MMAS)表现出独特的基因典型耐药谱,具有与适应和耐药机制相关的独特核心基因。字符串分析鉴定出42个独特的MAB核心基因,其中trub、prmC_2和aguA_2等11个基因互作得分最高。相比之下,MMAS有11个独特的核心基因,lgrD_4和rnc之间存在单一的相互作用。随后使用NCBI BLAST验证表明,只有fmt_2和aguA_2是MAB真正独特的。本研究为脓疡分枝杆菌在持续感染和再感染期间的基因组进化以及亚种间的遗传变异提供了新的见解。研究结果增强了对脓疡分枝杆菌流行病学的认识,并可能为治疗和感染控制提供信息。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Infection Genetics and Evolution
医学-传染病学
CiteScore
8.40
自引率
0.00%
发文量
215
审稿时长
82 days
期刊介绍:
(aka Journal of Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases -- MEEGID)
Infectious diseases constitute one of the main challenges to medical science in the coming century. The impressive development of molecular megatechnologies and of bioinformatics have greatly increased our knowledge of the evolution, transmission and pathogenicity of infectious diseases. Research has shown that host susceptibility to many infectious diseases has a genetic basis. Furthermore, much is now known on the molecular epidemiology, evolution and virulence of pathogenic agents, as well as their resistance to drugs, vaccines, and antibiotics. Equally, research on the genetics of disease vectors has greatly improved our understanding of their systematics, has increased our capacity to identify target populations for control or intervention, and has provided detailed information on the mechanisms of insecticide resistance.
However, the genetics and evolutionary biology of hosts, pathogens and vectors have tended to develop as three separate fields of research. This artificial compartmentalisation is of concern due to our growing appreciation of the strong co-evolutionary interactions among hosts, pathogens and vectors.
Infection, Genetics and Evolution and its companion congress [MEEGID](http://www.meegidconference.com/) (for Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases) are the main forum acting for the cross-fertilization between evolutionary science and biomedical research on infectious diseases.
Infection, Genetics and Evolution is the only journal that welcomes articles dealing with the genetics and evolutionary biology of hosts, pathogens and vectors, and coevolution processes among them in relation to infection and disease manifestation. All infectious models enter the scope of the journal, including pathogens of humans, animals and plants, either parasites, fungi, bacteria, viruses or prions. The journal welcomes articles dealing with genetics, population genetics, genomics, postgenomics, gene expression, evolutionary biology, population dynamics, mathematical modeling and bioinformatics. We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. Please click here for more information on our author services .
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


