Exome Sequencing in Saudi Arabian Pediatric Kidney Disease Single-Center Cohort
IF 5.7
2区 医学
Q1 UROLOGY & NEPHROLOGY
引用次数: 0
Abstract
Introduction
In pediatric patients, monogenic causes are a significant contributor to kidney disease, ranging from approximately 10% in congenital anomalies of the kidney and urinary tract (CAKUT) to about 55% in renal ciliopathies. Exome sequencing has revealed numerous disease-causing genes and pathogenic variants. Nevertheless, continuous efforts are crucial to expand the knowledge base of these variants to establish unequivocal diagnoses. In this study, we report exome sequencing data from a single Saudi Arabian center, aiming to explore potential founder effects and determine specific genotype-phenotype correlations based on clinical diagnoses and genetic ancestry.
Methods
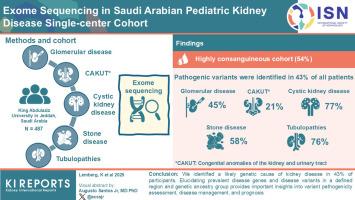
We consolidated 487 families with glomerular disease, CAKUT, cystic kidney disease, stone disease, and tubulopathies, recruited between 2007 and 2023 at King Abdulaziz University in Jeddah, Saudi Arabia. In these families, we performed exome sequencing and analyzed the data obtained for variants in established disease genes.
Results
In this highly consanguineous cohort (54%), 195 of 487 participants (40%) had glomerular disease, 160 of 487 (33%) had CAKUT, 44 of 487 (9%) had cystic kidney disease, 40 of 487 (8%) had stone disease, and 34 of 487 (7%) had tubulopathies. Pathogenic variants were identified in 45% of families with glomerular disease, 21% with CAKUT, 77% with cystic kidney disease, 58% with stone disease, and 76% of families with tubulopathy.
Conclusion
We identified a likely genetic cause of kidney disease in 43% of participants. Elucidating prevalent disease genes and disease variants in a defined region and genetic ancestry group provides important insights into variant pathogenicity assessment, disease management, and prognosis.
沙特阿拉伯儿童肾病单中心队列外显子组测序
在儿科患者中,单基因原因是肾脏疾病的重要因素,从约10%的先天性肾脏和尿路异常(CAKUT)到约55%的肾纤毛病。外显子组测序揭示了许多致病基因和致病变异。然而,持续的努力是至关重要的,以扩大这些变异的知识库,以建立明确的诊断。在这项研究中,我们报告了来自一个沙特阿拉伯中心的外显子组测序数据,旨在探索潜在的创始人效应,并根据临床诊断和遗传血统确定特定的基因型-表型相关性。方法:研究纳入了2007年至2023年在沙特阿拉伯吉达的阿卜杜勒阿齐兹国王大学(King Abdulaziz University)招募的487个肾小球疾病、CAKUT、囊性肾病、结石疾病和小管疾病家族。在这些家庭中,我们进行了外显子组测序,并分析了已确定疾病基因变异的数据。结果在这个高度近亲队列中(54%),487名参与者中有195名(40%)患有肾小球疾病,160名(33%)患有CAKUT, 44名(9%)患有囊肾疾病,40名(8%)患有结石疾病,34名(7%)患有小管疾病。45%的肾小球疾病家族、21%的CAKUT家族、77%的囊性肾病家族、58%的结石疾病家族和76%的小管病变家族都发现了致病变异。结论:我们在43%的参与者中发现了肾脏疾病的可能遗传原因。阐明特定地区和遗传祖先群体中的流行疾病基因和疾病变异为变异致病性评估、疾病管理和预后提供了重要见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Kidney International Reports
Medicine-Nephrology
CiteScore
7.70
自引率
3.30%
发文量
1578
审稿时长
8 weeks
期刊介绍:
Kidney International Reports, an official journal of the International Society of Nephrology, is a peer-reviewed, open access journal devoted to the publication of leading research and developments related to kidney disease. With the primary aim of contributing to improved care of patients with kidney disease, the journal will publish original clinical and select translational articles and educational content related to the pathogenesis, evaluation and management of acute and chronic kidney disease, end stage renal disease (including transplantation), acid-base, fluid and electrolyte disturbances and hypertension. Of particular interest are submissions related to clinical trials, epidemiology, systematic reviews (including meta-analyses) and outcomes research. The journal will also provide a platform for wider dissemination of national and regional guidelines as well as consensus meeting reports.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


