Exploring Feature Extraction Methods for Raman Spectroscopy: A Comparative Study
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
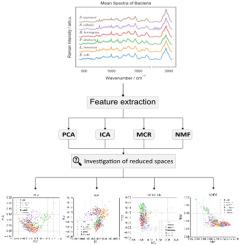
Raman spectroscopy is a robust, non-destructive analytical technique that offers detailed insights into the chemical composition, molecular structure, and interactions of materials. However, the high-dimensional and complex nature of Raman spectral data requires effective feature extraction methods to reduce data volume and improve analysis. Efficient feature extraction methods are essential to reduce dimensionality while preserving critical spectral information. This study investigates and compares four feature extraction techniques, Principal Component Analysis (PCA), Independent Component Analysis (ICA), Multivariate Curve Resolution (MCR), and Non-negative Matrix Factorization (NMF), in the context of Raman spectroscopy to assess their ability to reduce the dimensionality of high-dimensional spectral data while preserving critical chemical and biological information.Results
Using simulated datasets and real bacterial Raman spectra, we assessed how each method transformed high-dimensional Raman spectra into a lower-dimensional space, focusing on the structure of the reduced representations, their interpretability in terms of chemical and biological meaning, and their effectiveness in classification tasks. PCA and ICA effectively reduced dimensionality with minimal reconstruction errors, but they produced less interpretable features due to the orthogonality and independence constraints. In contrast, MCR and NMF generated chemically meaningful features and achieved classification performance comparable to that of PCA, even with fewer components.Significance
For the first time, this study provides a systematic and in-depth examination of the reduced feature spaces generated by each method, offering a comprehensive understanding of their structural properties, interpretability, and impact on classification performance. The results highlight MCR as a particularly promising method for feature extraction in Raman spectroscopy. Its ability to produce chemically interpretable features and incorporate physicochemical constraints offers a key advantage over conventional techniques such as PCA. These characteristics make MCR especially suitable for analyzing complex Raman spectral data, where both classification accuracy and interpretability are crucial.
探索拉曼光谱特征提取方法:比较研究
draman光谱是一种强大的、非破坏性的分析技术,可以提供对化学成分、分子结构和材料相互作用的详细见解。然而,拉曼光谱数据的高维性和复杂性需要有效的特征提取方法来减少数据量,提高分析能力。有效的特征提取方法对于降低维数的同时保留关键的光谱信息至关重要。本研究对拉曼光谱中的主成分分析(PCA)、独立成分分析(ICA)、多元曲线分辨率(MCR)和非负矩阵分解(NMF)四种特征提取技术进行了研究和比较,以评估它们在降低高维光谱数据维数的同时保留关键化学和生物信息的能力。结果利用模拟数据集和真实细菌拉曼光谱,我们评估了每种方法如何将高维拉曼光谱转换为低维空间,重点关注了降维表征的结构、化学和生物意义的可解释性以及它们在分类任务中的有效性。PCA和ICA在重构误差最小的情况下可以有效地降维,但由于正交性和独立性的限制,它们产生的可解释特征较少。相比之下,MCR和NMF生成了化学上有意义的特征,即使成分更少,分类性能也与PCA相当。本研究首次对每种方法生成的约简特征空间进行了系统和深入的研究,全面了解了它们的结构特性、可解释性以及对分类性能的影响。结果表明MCR是一种特别有前途的拉曼光谱特征提取方法。与PCA等传统技术相比,它能够产生化学可解释特征并结合物理化学约束,这是一个关键优势。这些特点使得MCR特别适合分析复杂的拉曼光谱数据,其中分类精度和可解释性都是至关重要的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


