Genetic differentiation of Pisang Awak subvarieties and genetic variation among ‘Mali-Ong’ plantlets in Thailand using RAPD and SRAP markers
IF 2.8
Q3 Biochemistry, Genetics and Molecular Biology
Journal of Genetic Engineering and Biotechnology
Pub Date : 2025-10-03
DOI:10.1016/j.jgeb.2025.100577
引用次数: 0
Abstract
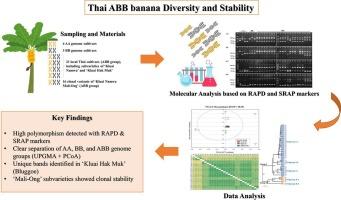
Banana (Musa spp.) is a globally important fruit crop, with most cultivated varieties originating from hybridizations between M. acuminata (A genome) and M. balbisiana (B genome). Triploid ABB hybrids, carrying two B-genome and one A-genome sets, are valued for their stress tolerance and adaptability. In Thailand, ‘Kluai Namwa’ (Pisang Awak) is the most widely cultivated ABB cultivar, but it shows considerable phenotypic variation across subvarieties. Because morphological classification is often unreliable, molecular tools are needed to assess genetic identity and diversity. A total of 28 Thai banana genotypes, representing the AA, BB, and ABB genome groups, were analyzed using RAPD and SRAP markers. RAPD produced 109 bands with 93.6 % polymorphism, while SRAP generated 278 bands with 92.5 % polymorphism, indicating substantial genetic variation. The mean polymorphic information content was 0.22 for RAPD and 0.24 for SRAP, confirming the discriminatory power of both marker systems. UPGMA clustering separated the genotypes into two major clusters corresponding to A- and B-genome contributions, a structure further supported by PCoA. Distinctive bands, such as RAPD primer S7 (2.50 kb) and SRAP combination Me6/Em8 (0.80 kb), specifically identified ‘Kluai Hak Muk’ cooking bananas, demonstrating the potential of these markers for cultivar authentication. The genetic stability of 16 ‘Kluai Namwa Mali-Ong’ plantlets from different locations was also evaluated. Results revealed high clonal uniformity (mean similarity = 0.858) with only minor variation, likely reflecting localized cultivation practices. Overall, RAPD and SRAP markers proved effective for genome identification, diversity assessment, and clonal stability monitoring in Thai bananas. These tools will support banana breeding, germplasm conservation, and reliable authentication of high-value cultivars.
利用RAPD和SRAP标记对泰国“麻荔ong”植株的遗传变异及Pisang Awak亚种的遗传分化
香蕉(Musa spp.)是一种全球重要的水果作物,大多数栽培品种起源于M. acuminata (a基因组)和M. balbisiana (B基因组)的杂交。三倍体ABB杂交种具有2组b基因组和1组a基因组,具有较强的抗逆性和适应性。在泰国,“Kluai Namwa”(Pisang Awak)是种植最广泛的ABB品种,但它在亚种之间表现出相当大的表型差异。由于形态学分类往往是不可靠的,需要分子工具来评估遗传身份和多样性。利用RAPD和SRAP标记分析了泰国香蕉共28个基因型,分别代表AA、BB和ABB基因组群。RAPD产生109个条带,多态性为93.6%,SRAP产生278个条带,多态性为92.5%,遗传变异较大。RAPD的平均多态性信息含量为0.22,SRAP的平均多态性信息含量为0.24,证实了两种标记系统的区分能力。UPGMA聚类将基因型分为A和b基因组贡献的两个主要簇,PCoA进一步支持了这一结构。RAPD引物S7 (2.50 kb)和SRAP组合Me6/Em8 (0.80 kb)等独特的条带特异性地鉴定了“Kluai Hak Muk”烹饪香蕉,表明这些标记在品种鉴定方面具有潜力。并对不同产地的16个‘Kluai Namwa Mali-Ong’植株的遗传稳定性进行了评价。结果表明,克隆均一性较高(平均相似度= 0.858),变异较小,可能反映了栽培方式的局限性。总的来说,RAPD和SRAP标记在泰国香蕉的基因组鉴定、多样性评估和克隆稳定性监测方面是有效的。这些工具将为香蕉育种、种质资源保存和高价值品种的可靠鉴定提供支持。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Genetic Engineering and Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
5.70
自引率
5.70%
发文量
159
审稿时长
16 weeks
期刊介绍:
Journal of genetic engineering and biotechnology is devoted to rapid publication of full-length research papers that leads to significant contribution in advancing knowledge in genetic engineering and biotechnology and provide novel perspectives in this research area. JGEB includes all major themes related to genetic engineering and recombinant DNA. The area of interest of JGEB includes but not restricted to: •Plant genetics •Animal genetics •Bacterial enzymes •Agricultural Biotechnology, •Biochemistry, •Biophysics, •Bioinformatics, •Environmental Biotechnology, •Industrial Biotechnology, •Microbial biotechnology, •Medical Biotechnology, •Bioenergy, Biosafety, •Biosecurity, •Bioethics, •GMOS, •Genomic, •Proteomic JGEB accepts
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


