Potential small-molecule compounds and targets for Alzheimer's disease: Integrating bioinformatics analysis and in vitro verification
引用次数: 0
Abstract
Objective
Alzheimer's disease (AD) is a common neurodegenerative disorder characterized by progressive memory decline and cognitive dysfunction. The specific pathogenesis of AD remains unclear. This study aimed to explore the crucial genes and therapeutic small-molecule compounds in AD via integrated bioinformatics analysis, molecular docking, and in vitro verification.
Methods
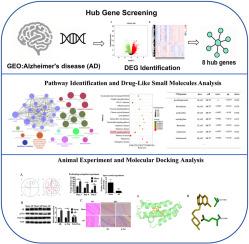
The gene dataset GSE122063, including 12 samples from patients with AD and 11 non-demented control samples, was downloaded from the Gene Expression Omnibus (GEO) database. The online tool GEO2R was used to analyze differentially expressed genes (DEGs). Functional enrichment analysis of the DEGs was performed using DAVID and ClueGo databases. A protein-protein interaction network was constructed using the STRING database and visualized in Cytoscape. Potential small-molecule compounds for AD therapy were screened using the Connectivity map database. The crucial genes in a rat model of AD were confirmed by RT-PCR. Molecular docking of the screened crucial genes and small-molecule compounds was further performed to identify potential therapeutic drugs for AD.
Results
A total of 1145 DEGs were identified, which were enriched in intracellular protein transport, cell cycle, establishment of protein localization to membrane, and so on. Eight hub genes, including RPS29, CREBBP, ANAPC10, ANAPC4, MAGOHB, TCEB2, RPL10A, and SEC61A1, were identified in the protein-protein interaction network. The MAPK signaling pathway was closely related to AD. Furthermore, increased expression of CREBBP was confirmed in the rat model of AD, and molecular docking revealed that CREBBP exhibited the strongest binding affinity with prochlorperazine.
Conclusion
CREBBP was identified as a crucial hub gene and might serve as a potential target for AD. Prochlorperazine, which exhibited strong binding to CREBBP, showed potential as a therapeutic drug in AD.
阿尔茨海默病潜在的小分子化合物和靶点:整合生物信息学分析和体外验证
目的阿尔茨海默病(AD)是一种常见的神经退行性疾病,以进行性记忆衰退和认知功能障碍为特征。阿尔茨海默病的具体发病机制尚不清楚。本研究旨在通过综合生物信息学分析、分子对接和体外验证,探索AD的关键基因和治疗性小分子化合物。方法从gene Expression Omnibus (GEO)数据库中下载基因数据集GSE122063,包括12例AD患者样本和11例非痴呆对照样本。使用在线工具GEO2R分析差异表达基因(DEGs)。使用DAVID和ClueGo数据库进行deg的功能富集分析。利用STRING数据库构建蛋白-蛋白相互作用网络,并在Cytoscape中可视化。使用Connectivity map数据库筛选潜在的用于阿尔茨海默病治疗的小分子化合物。RT-PCR证实了大鼠AD模型中的关键基因。筛选的关键基因和小分子化合物的分子对接进一步确定潜在的阿尔茨海默病治疗药物。结果共鉴定出1145个基因,富集于细胞内蛋白转运、细胞周期、蛋白向膜定位等方面。在蛋白相互作用网络中鉴定出8个枢纽基因,包括RPS29、CREBBP、ANAPC10、ANAPC4、MAGOHB、TCEB2、RPL10A和SEC61A1。MAPK信号通路与AD密切相关。此外,在AD大鼠模型中证实CREBBP表达增加,分子对接显示CREBBP与丙氯拉嗪的结合亲和力最强。结论crebbp是一个重要的中枢基因,可能是AD的潜在靶点。丙氯哌嗪与CREBBP结合较强,具有治疗AD的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


