Literature-based explainable machine learning models for predicting pathogen and antibiotic resistance gene loads from animal manure
IF 4
4区 环境科学与生态学
Q2 ENVIRONMENTAL SCIENCES
引用次数: 0
Abstract
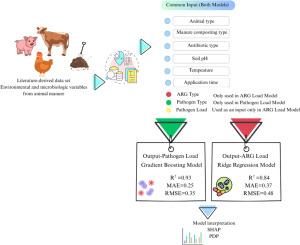
The use of animal manure (cattle, pigs, poultry, and sheep) in agriculture offers significant advantages such as increasing soil fertility and reducing the use of chemical fertilizers. However, this application also brings about serious environmental and public health problems due to the risk of microbial contaminants such as pathogenic microorganisms and antibiotic resistance genes (ARGs) spreading into the environment. In order to assess this dual risk, we developed a machine learning (ML) framework capable of simultaneously predicting pathogen load and ARG levels. The dataset contains 223 records systematically collected from 54 scientific studies published between 2015 and 2024. Six regression models were compared; Gradient Boosting algorithm (R2 = 0.93) for pathogen load and Ridge Regression algorithm (R2 = 0.84) for ARG level showed the highest accuracy performance. Model generalizability was tested with 5- and 10-fold cross-validation; low overfitting risk was confirmed by learning curves and residual analysis, specifically for the final selected models (Gradient Boosting for pathogen load and Ridge Regression for ARG level), while other models such as Decision Tree showed clear signs of overfitting and were therefore excluded from further analysis. The transparency of model decisions was examined with SHapley Additive exPlanations (SHAP) analyses; “application period”, “ARG type” and “fertilizer type” were highlighted as determining variables. In addition, Partial Dependence Plot (PDP) analyses revealed the marginal effects of environmental and operational factors on target variables in a biologically meaningful way. This integrated modelling approach contributes to the optimization of sustainable fertilization strategies and the development of environmental-health policies.
基于文献的可解释机器学习模型,用于预测动物粪便中的病原体和抗生素抗性基因负荷
在农业中使用动物粪便(牛、猪、家禽和羊)具有显著的优势,如提高土壤肥力和减少化肥的使用。然而,由于病原微生物和抗生素耐药基因(ARGs)等微生物污染物扩散到环境中的风险,这种应用也带来了严重的环境和公共卫生问题。为了评估这种双重风险,我们开发了一个能够同时预测病原体负荷和ARG水平的机器学习(ML)框架。该数据集包含223条记录,系统收集了2015年至2024年间发表的54项科学研究。比较6种回归模型;病原菌负荷梯度增强算法(R2 = 0.93)和ARG水平岭回归算法(R2 = 0.84)的准确率最高。用5倍和10倍交叉验证检验模型的可推广性;学习曲线和残差分析证实了低过拟合风险,特别是对于最终选择的模型(用于病原体负荷的梯度增强和用于ARG水平的Ridge回归),而其他模型(如Decision Tree)显示出明显的过拟合迹象,因此被排除在进一步分析之外。采用SHapley加性解释(SHAP)分析检验模型决策的透明度;“施肥期”、“ARG类型”和“肥料类型”是决定变量。此外,偏相关图(PDP)分析以生物学意义的方式揭示了环境和操作因素对目标变量的边际效应。这种综合建模方法有助于优化可持续施肥战略和制定环境卫生政策。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Microbial Risk Analysis
Medicine-Microbiology (medical)
CiteScore
5.70
自引率
7.10%
发文量
28
审稿时长
52 days
期刊介绍:
The journal Microbial Risk Analysis accepts articles dealing with the study of risk analysis applied to microbial hazards. Manuscripts should at least cover any of the components of risk assessment (risk characterization, exposure assessment, etc.), risk management and/or risk communication in any microbiology field (clinical, environmental, food, veterinary, etc.). This journal also accepts article dealing with predictive microbiology, quantitative microbial ecology, mathematical modeling, risk studies applied to microbial ecology, quantitative microbiology for epidemiological studies, statistical methods applied to microbiology, and laws and regulatory policies aimed at lessening the risk of microbial hazards. Work focusing on risk studies of viruses, parasites, microbial toxins, antimicrobial resistant organisms, genetically modified organisms (GMOs), and recombinant DNA products are also acceptable.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


