Structural basis for DCAF2 as a novel E3 ligase for PROTAC-mediated targeted protein degradation
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
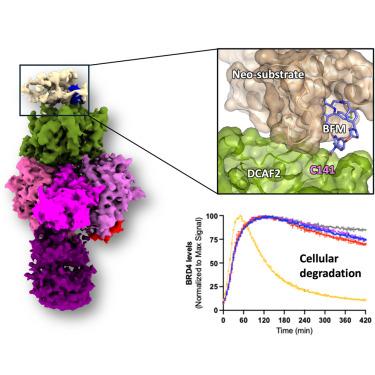
Targeted protein degradation (TPD) leverages the ubiquitin-proteasome system to eliminate disease-causing proteins via E3 ligases. To date, the field is limited to utilizing a few of the over 600 human E3 ligases. To expand this repertoire, we conducted structural and functional validation of DDB1 (Damage-specific DNA binding protein 1) and Cullin-associated factor (DCAF)2 (DTL/CDT2), a Cullin4-RING ligase substrate adaptor implicated in DNA damage response and cancer, as a novel E3 for TPD. Cryoelectron microscopy (cryo-EM) structures of the DCAF2:DDB1:DDA1 complex (3.3 Å), a ligand bound complex (3.1 Å), and a ternary complex with a covalent proteolysis-targeting chimera (PROTAC) and BRD4 (3.4 Å) reveal PROTAC-mediated substrate recruitment. Using covalent bifunctional tool compounds engaging residue C141 in the WD40 domain, we demonstrate robust ubiquitination in biochemical assays and cellular TPD using the COFFEE (covalent functionalization followed by E3 electroporation) method. These findings position DCAF2 as a promising E3 adaptor for PROTAC strategies and identify C141 as a relevant site for future PROTAC discovery.
DCAF2作为protac介导的靶向蛋白降解的新型E3连接酶的结构基础
靶向蛋白降解(TPD)利用泛素-蛋白酶体系统通过E3连接酶消除致病蛋白。迄今为止,该领域仅限于利用600多个人类E3连接中的几个。为了扩大这一范围,我们对DDB1(损伤特异性DNA结合蛋白1)和cullin相关因子(DCAF)2 (DTL/CDT2)进行了结构和功能验证,Cullin4-RING连接酶底物适配器参与DNA损伤反应和癌症,作为TPD的新E3。DCAF2:DDB1:DDA1配合物(3.3 Å)、配体结合配合物(3.1 Å)以及具有共价蛋白水解靶向嵌合体(PROTAC)和BRD4 (3.4 Å)的三元配合物的冷冻电镜(cro - em)结构揭示了PROTAC介导的底物募集。我们使用含有WD40结构域C141残基的共价双功能工具化合物,在生化分析和使用COFFEE(共价功能化后E3电穿孔)方法的细胞TPD中证明了强大的泛素化作用。这些发现将DCAF2定位为PROTAC策略的有前途的E3适配器,并确定C141是未来发现PROTAC的相关位点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


