Connecting chemical and protein sequence space to predict biocatalytic reactions
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
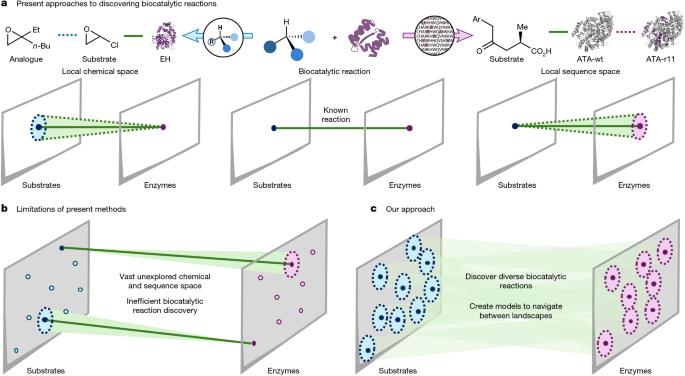
The application of biocatalysis in synthesis has the potential to offer streamlined routes towards target molecules1, tunable catalyst-controlled selectivity2, as well as processes with improved sustainability3. Despite these advantages, biocatalysis is often a high-risk strategy to implement, as identifying an enzyme capable of performing chemistry on a specific intermediate required for a synthesis can be a roadblock that requires extensive screening of enzymes and protein engineering to overcome4. Strategies for predicting which enzyme and small molecule are compatible have been hindered by the lack of well-studied biocatalytic reaction datasets5. The underexploration of connections between chemical and protein sequence space constrains navigation between these two landscapes. Here we report a two-phase effort relying on high-throughput experimentation to populate connections between productive substrate and enzyme pairs and the subsequent development of a tool, CATNIP, for predicting compatible α-ketoglutarate (α-KG)/Fe(ii)-dependent enzymes for a given substrate or, conversely, for ranking potential substrates for a given α-KG/Fe(ii)-dependent enzyme sequence. We anticipate that our approach can be readily expanded to further enzyme and transformation classes and will derisk the investigation and application of biocatalytic methods. A two-phase machine-learning-based tool making use of high-throughput experimentation is introduced to examine the connections between chemical and protein sequence space and predict productive biocatalytic reactions among substrate and enzyme pairs.
连接化学和蛋白质序列空间以预测生物催化反应
生物催化在合成中的应用有可能提供通向目标分子的流线型路线1,可调节的催化剂控制的选择性2,以及提高可持续性的过程3。尽管有这些优势,生物催化通常是一种高风险的策略,因为识别一种能够对合成所需的特定中间体进行化学反应的酶可能是一个障碍,需要大量的酶筛选和蛋白质工程才能克服。由于缺乏经过充分研究的生物催化反应数据集,预测哪些酶和小分子相容的策略受到了阻碍。对化学和蛋白质序列空间之间联系的探索不足限制了这两种景观之间的导航。在这里,我们报告了两阶段的努力,依靠高通量实验来建立生产底物和酶对之间的联系,以及随后开发的工具CATNIP,用于预测给定底物的相容α-酮戊二酸(α-KG)/Fe(ii)依赖性酶,或者相反,用于对给定α-KG/Fe(ii)依赖性酶序列的潜在底物进行排序。我们预计我们的方法可以很容易地扩展到进一步的酶和转化类,并将冒险的研究和应用生物催化方法。介绍了一种基于两阶段机器学习的工具,该工具利用高通量实验来检查化学和蛋白质序列空间之间的联系,并预测底物和酶对之间的生产性生物催化反应。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


