Sequencing-free whole-genome spatial transcriptomics at single-molecule resolution
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Recent breakthroughs in spatial transcriptomics technologies have enhanced our understanding of diverse cellular identities, spatial organizations, and functions. Yet existing spatial transcriptomics tools are still limited in either transcriptomic coverage or spatial resolution, hindering unbiased, hypothesis-free transcriptomic analyses at high spatial resolution. Here, we develop reverse-padlock amplicon-encoding fluorescence in situ hybridization (RAEFISH), an image-based spatial transcriptomics method with whole-genome coverage and single-molecule resolution in intact tissues. We demonstrate the spatial profiling of transcripts from 23,000 human or 22,000 mouse genes in single cells and tissue sections. Our analyses reveal transcript-specific subcellular localization, cell-type-specific and cell-type-invariant zonation-dependent transcriptomes, and gene programs underlying preferential cell-cell interactions. Finally, we further develop our technology for the direct spatial readout of guide RNAs (gRNAs) in an image-based, high-content CRISPR screen. Overall, these developments offer a broadly applicable technology that enables high-coverage, high-resolution spatial profiling of both long and short, native and engineered RNAs in many biomedical contexts.
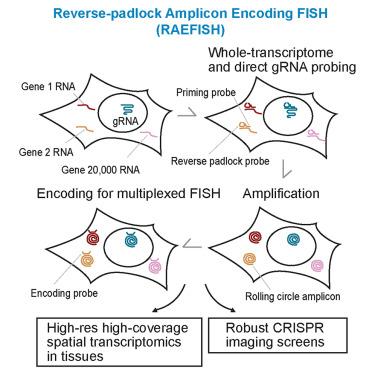
单分子分辨率无测序全基因组空间转录组学
空间转录组学技术的最新突破增强了我们对不同细胞身份、空间组织和功能的理解。然而,现有的空间转录组学工具在转录组覆盖范围或空间分辨率方面仍然有限,阻碍了高空间分辨率下无偏见、无假设的转录组学分析。在这里,我们开发了反向挂锁扩增子编码荧光原位杂交(RAEFISH),这是一种在完整组织中具有全基因组覆盖和单分子分辨率的基于图像的空间转录组学方法。我们在单个细胞和组织切片中展示了23,000个人类或22,000个小鼠基因的转录本的空间分析。我们的分析揭示了转录特异性亚细胞定位,细胞类型特异性和细胞类型不变的区依赖转录组,以及潜在的优先细胞-细胞相互作用的基因程序。最后,我们进一步开发了在基于图像的高含量CRISPR屏幕中直接空间读出引导rna (grna)的技术。总的来说,这些发展提供了一种广泛适用的技术,可以在许多生物医学背景下对长rna和短rna、天然rna和工程rna进行高覆盖、高分辨率的空间分析。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


