Proteasome-associated autoinflammatory syndromes: The impact of mutations in proteasome subunits on particle assembly, structure, and activity
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Single point mutations in proteasome subunits can cause severe autoinflammatory syndromes. By still largely unknown mechanisms, some of these disease-associated mutations impair normal proteasome function and induce the production of pro-inflammatory cytokines, thereby leading to systemic inflammations. In order to obtain more insights on why and how the mutations T3M and G128V in the immunoproteasome subunit β5i trigger such deleterious effects, we created the respective yeast mutants and characterized their phenotypes with special emphasis on proteasome structure and activity. X-ray crystallographic data revealed that the mutation T3M influences structure and flexibility of the proteasomal substrate-binding channel with moderate impairment of proteasome biogenesis, whereas the amino acid substitution G128V causes larger structural rearrangements that severely disturb particle assembly and maturation. The obtained results provide a deeper understanding of how single point mutations can affect proteasome subunit structure as well as particle biogenesis and ultimately cause chronic inflammatory diseases.
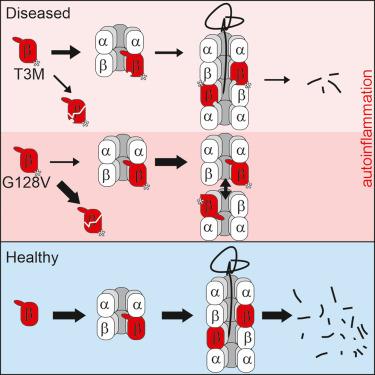
蛋白酶体相关的自身炎症综合征:蛋白酶体亚基突变对颗粒组装、结构和活性的影响
蛋白酶体亚基的单点突变可引起严重的自身炎症综合征。其中一些疾病相关的突变破坏了正常的蛋白酶体功能,诱导促炎细胞因子的产生,从而导致全身性炎症,其机制在很大程度上仍然未知。为了更深入地了解免疫蛋白酶体亚基β5i中的突变T3M和G128V为何以及如何引发这种有害效应,我们创建了各自的酵母突变体,并对其表型进行了表征,特别强调蛋白酶体的结构和活性。x射线晶体学数据显示,突变T3M影响蛋白酶体底物结合通道的结构和灵活性,中度损害蛋白酶体的生物发生,而氨基酸取代G128V导致较大的结构重排,严重扰乱颗粒组装和成熟。获得的结果为单点突变如何影响蛋白酶体亚基结构以及颗粒生物发生并最终导致慢性炎症性疾病提供了更深入的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


