Sequential denaturation and protein precipitation assay (SDPP) improves the sensitivity for ligand target identification at the proteome level
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
Proteins can be denatured and precipitated under different denaturation conditions, and ligand bound proteins are more resistant to denaturing induced precipitation. Consequently, ligand target proteins can be identified by measuring solubility shifts. Nowadays, numerous methods have been developed to screen ligand target proteins based on different denaturation mechanisms. However, due to the different responses of proteins to different denaturation conditions, there is a significant complementary between these methods. Direct pooling of the denatured supernatants from different denaturation strategies can utilize this complementary, however, the pooling procedure inevitably averages the ligand-induced solubility shift across conditions, significantly compromising target identification sensitivity.
Results
To avoid pooling-induced signal compression and enhance identification sensitivity, we developed a novel solubility shift-based approach, termed Sequential Denaturation and Protein Precipitation assay (SDPP). In this strategy, multi-step sequential denaturation of individual sample was conducted, which greatly improves the identification sensitivity. We evaluated the performance of this method by screening the target proteins of Staurosporine, SDPP with thermal treatment followed by organic solvent treatment (TEMP-SL) performs better than other sequential denaturation and one-step denaturation approach. The number of kinase targets identified by SDPP (TEMP-SL) increased by 54 %, 38 %, 48 % and 21 %, respectively compared with the TEMP, SL, and pH experiments and IPSSA. We further applied this approach to identify the target proteins of endogenous metabolite cAMP. Except for revealing the known cAMP-PRKAR1A/PRKAR1B interaction, we also observed that the solubility shift of the C2orf88 responds to the cAMP binding.
Significance
This method conducts sequential denaturation on individual samples to induce stepwise protein precipitation, preserving and progressively accumulating the solubility shifts generated at each denaturation step. This cumulative effect amplifies the overall detectable solubility shift signal and greatly improves identification sensitivity, demonstrating great potential as an efficient and high-throughput strategy for proteome-wide ligand target identification.
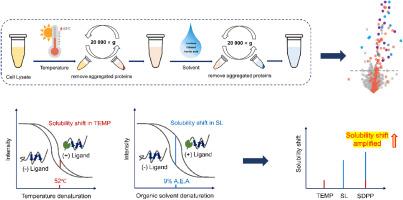
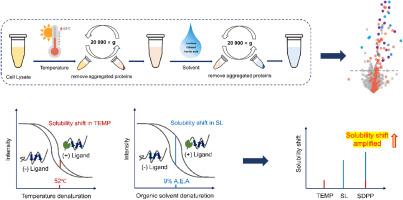
序列变性和蛋白质沉淀试验(SDPP)提高了在蛋白质组水平上对配体靶标识别的灵敏度
蛋白质在不同的变性条件下会发生变性和沉淀,配体结合蛋白对变性诱导沉淀的抵抗力更强。因此,配体靶蛋白可以通过测量溶解度变化来识别。目前,基于不同的变性机制,已经开发了许多方法来筛选配体靶蛋白。然而,由于蛋白质对不同变性条件的反应不同,这些方法之间存在显著的互补性。来自不同变性策略的变性上清液的直接池化可以利用这种互补,然而,池化过程不可避免地平均了配体诱导的溶解度在不同条件下的变化,显著损害了目标识别的敏感性。为了避免池诱导的信号压缩并提高识别灵敏度,我们开发了一种新的基于溶解度位移的方法,称为顺序变性和蛋白质沉淀测定(SDPP)。该策略对单个样品进行多步顺序变性,大大提高了识别灵敏度。通过对Staurosporine的靶蛋白进行筛选,结果表明,SDPP经过热处理后再进行有机溶剂处理(TEMP-SL)的效果优于其他顺序变性和一步变性方法。与TEMP、SL、pH实验和IPSSA相比,SDPP (TEMP-SL)鉴定出的激酶靶点数量分别增加了54%、38%、48%和21%。我们进一步应用该方法鉴定内源性代谢物cAMP的靶蛋白。除了揭示已知的cAMP- prkar1a /PRKAR1B相互作用外,我们还观察到C2orf88的溶解度变化响应cAMP结合。该方法对单个样品进行顺序变性,诱导蛋白质逐步沉淀,保存并逐步积累每个变性步骤产生的溶解度变化。这种累积效应放大了整体可检测的溶解度偏移信号,极大地提高了识别灵敏度,显示出作为蛋白质组级配体靶标识别的高效和高通量策略的巨大潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


