Construction of a circRNA-miRNA-mRNA ceRNA regulatory network identifies RNAs and genes linked to human ovarian clear cell carcinoma
IF 3
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Background
Ovarian clear cell carcinoma (OCCC) is an aggressive epithelial ovarian cancer subtype that prevalent in East Asia. Competing endogenous RNA (ceRNA) networks involving circular RNAs (circRNAs) and microRNAs (miRNAs) represent underexplored regulatory layers in OCCC pathogenesis.
Methods
Multi-omics integration of circRNA (GSE271851 and GSE266248) and miRNA (GSE230956 and GSE200852) datasets from GEO was performed using DESeq2 and limma tools (|log2FC|≥1, p < 0.05). Experimentally validated circRNA-miRNA interactions were predicted using circBank 2.0, Circular RNA Interactome, and Starbase v3.0. ceRNA networks were constructed using inverse correlation principles and visualized in Cytoscape. Immune associations were assessed using TIMER2.0. Prognostic and expression validations were performed using the Kaplan-Meier Plotter and HPA databases, respectively.
Results
Three core circRNAs (hsa_circ_0002822, hsa_circ_0003641 and hsa_circ_0030509) were identified as being dysregulated across OCCC models. Their miRNA sponging activity formed a 65-node ceRNA network involving 15 miRNAs (e.g., miR-143–3p and miR-182–5p) and 47 mRNAs. Six targets (BRCA1, KRAS, MSH6, SMARCA4, SRC, and TSC1) exhibited significant correlations with immune infiltration (CD8+ T cells, Tregs, and MDSCs). High expression of BRCA1, KRAS, and MSH6 predicted poor survival, with protein-level upregulation confirmed in OCCC tissues.
Conclusion
This study delineates a circRNA-driven ceRNA network in OCCC, and identifies BRCA1, KRAS, and MSH6 as multi-omic biomarkers with immune-modulatory roles. These findings provide a basis for the development of RNA-targeted therapies against this ovarian cancer subtype.
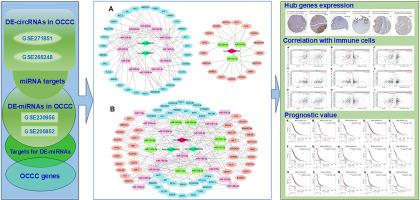
构建circRNA-miRNA-mRNA ceRNA调控网络鉴定与人卵巢透明细胞癌相关的rna和基因
背景:卵巢透明细胞癌(OCCC)是一种侵袭性上皮性卵巢癌亚型,常见于东亚地区。涉及环状RNA (circRNAs)和microRNAs (miRNAs)的竞争性内源性RNA (ceRNA)网络代表了OCCC发病机制中未被充分探索的调控层。方法:使用DESeq2和limma工具(|log2FC|≥1)对GEO的circRNA (GSE271851和GSE266248)和miRNA (GSE230956和GSE200852)数据集进行多组学整合。结果:三个核心circRNA (hsa_circ_0002822, hsa_circ_0003641和hsa_circ_0030509)在OCCC模型中被鉴定为失调。它们的miRNA海绵活性形成了一个65节点的ceRNA网络,涉及15个miRNA(例如miR-143-3p和miR-182-5p)和47个mrna。6个靶点(BRCA1、KRAS、MSH6、SMARCA4、SRC和TSC1)与免疫浸润(CD8+ T细胞、Tregs和MDSCs)有显著相关性。BRCA1、KRAS和MSH6的高表达预示着较差的生存率,在OCCC组织中证实了蛋白水平上调。结论:本研究描述了OCCC中circrna驱动的ceRNA网络,并确定了BRCA1、KRAS和MSH6作为具有免疫调节作用的多组学生物标志物。这些发现为开发针对这种卵巢癌亚型的rna靶向治疗提供了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Archives of biochemistry and biophysics
生物-生化与分子生物学
CiteScore
7.40
自引率
0.00%
发文量
245
审稿时长
26 days
期刊介绍:
Archives of Biochemistry and Biophysics publishes quality original articles and reviews in the developing areas of biochemistry and biophysics.
Research Areas Include:
• Enzyme and protein structure, function, regulation. Folding, turnover, and post-translational processing
• Biological oxidations, free radical reactions, redox signaling, oxygenases, P450 reactions
• Signal transduction, receptors, membrane transport, intracellular signals. Cellular and integrated metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


