DeepSES: Learning solvent-excluded surfaces via neural signed distance fields
IF 2.8
4区 计算机科学
Q2 COMPUTER SCIENCE, SOFTWARE ENGINEERING
引用次数: 0
Abstract
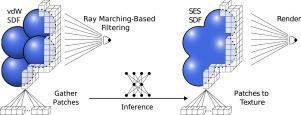
The solvent-excluded surface (SES) is essential for revealing molecular shape and solvent accessibility in applications such as molecular modeling, drug discovery, and protein folding. Its signed distance field (SDF) delivers a continuous, differentiable surface representation that enables efficient rendering, analysis, and interaction in volumetric visualization frameworks. However, analytic methods that compute the SDF of the SES cannot run at interactive rates on large biomolecular complexes, and grid-based methods tend to result in significant approximation errors, depending on molecular size and grid resolution. We address these limitations with DeepSES, a neural inference pipeline that predicts the SES SDF directly from the computationally simpler van der Waals (vdW) SDF on a fixed high-resolution grid. By employing an adaptive volume-filtering scheme that directs processing only to visible regions near the molecular surface, DeepSES yields interactive frame rates irrespective of molecule size. By offering multiple network configurations, DeepSES enables practitioners to balance inference time against prediction accuracy. In benchmarks on molecules ranging from one thousand to nearly four million atoms, our fastest configuration achieves real-time frame rates with a sub-angstrom mean error, while our highest-accuracy variant sustains interactive performance and outperforms state-of-the-art methods in terms of surface quality. By replacing costly algorithmic solvers with selective neural prediction, DeepSES provides a scalable, high-resolution solution for interactive biomolecular visualization.
DeepSES:通过神经符号距离场学习溶剂排除表面
溶剂排除表面(SES)在分子建模、药物发现和蛋白质折叠等应用中对于揭示分子形状和溶剂可及性至关重要。它的符号距离域(SDF)提供了一个连续的、可微的表面表示,在体积可视化框架中实现了高效的渲染、分析和交互。然而,计算SES的SDF的分析方法不能在大型生物分子复合物上以交互速率运行,并且基于网格的方法往往会导致显着的近似误差,这取决于分子大小和网格分辨率。我们使用DeepSES解决了这些限制,DeepSES是一种神经推理管道,可以直接从固定高分辨率网格上计算更简单的范德瓦尔斯(vdW) SDF预测SES SDF。通过采用自适应体积滤波方案,只对分子表面附近的可见区域进行处理,DeepSES产生的交互帧率与分子大小无关。通过提供多种网络配置,DeepSES使从业者能够平衡推理时间和预测准确性。在从1000到近400万个原子的分子基准测试中,我们最快的配置实现了亚埃平均误差的实时帧率,而我们最高精度的变体保持了交互性能,并在表面质量方面优于最先进的方法。通过用选择性神经预测取代昂贵的算法求解器,DeepSES为交互式生物分子可视化提供了可扩展的高分辨率解决方案。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers & Graphics-Uk
工程技术-计算机:软件工程
CiteScore
5.30
自引率
12.00%
发文量
173
审稿时长
38 days
期刊介绍:
Computers & Graphics is dedicated to disseminate information on research and applications of computer graphics (CG) techniques. The journal encourages articles on:
1. Research and applications of interactive computer graphics. We are particularly interested in novel interaction techniques and applications of CG to problem domains.
2. State-of-the-art papers on late-breaking, cutting-edge research on CG.
3. Information on innovative uses of graphics principles and technologies.
4. Tutorial papers on both teaching CG principles and innovative uses of CG in education.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


