PowderBot: An automated device for decision-making in crop breeding programs based on DNA extraction from seed powder
IF 2.1
Q3 ENGINEERING, ELECTRICAL & ELECTRONIC
引用次数: 0
Abstract
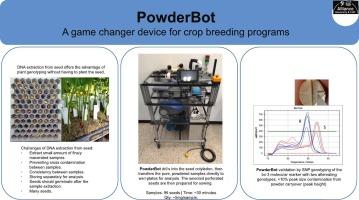
This paper presents the design, construction, operation, and evaluation of PowderBot, a purpose-built, open-source, low-cost machine (∼US$ 5000) that automates DNA extraction from ungerminated seeds. The device drills into the seed cotyledon, where the genetic information of the prospective plant is stored. It then transfers the pure, powdered samples directly to well-plates for analysis. This reduces time and other research resources and can accelerate crop varietal improvement, ultimately contributing to more efficient and successful crop breeding programs. At CIAT́s campus, we have validated the method for obtaining seed-tissue material from common beans for DNA extraction and subsequently genotyping agronomically interesting lines, using the bc-3 molecular marker. Three genotyping trials were carried out using this method, which generated consistent results, regardless of the number of perforations made to the seed. This leads us to infer that the method works effectively and can be applied for marker assisted selection (MAS) in bean and other crop breeding programs. Finally, germination and vigor tests indicated the sampling process did not significantly compromise perforated seed germination rate, physiological quality or viability.
PowderBot:一种基于从种子粉中提取DNA的作物育种计划决策的自动化设备
本文介绍了PowderBot的设计、构建、操作和评估,这是一种专用的、开源的、低成本的机器(约5000美元),可以自动从未发芽的种子中提取DNA。该装置钻入种子子叶,那里储存着未来植物的遗传信息。然后将纯净的粉末状样品直接转移到孔板上进行分析。这减少了时间和其他研究资源,可以加速作物品种改良,最终有助于更有效和成功的作物育种计划。在CIAT的校园,我们已经验证了从普通豆类中获得种子组织材料的方法,用于DNA提取,随后使用bc-3分子标记对农学上感兴趣的品系进行基因分型。使用这种方法进行了三次基因分型试验,无论对种子穿孔的数量如何,结果都一致。这使我们推断该方法是有效的,可以应用于豆类和其他作物育种计划中的标记辅助选择(MAS)。最后,萌发和活力测试表明,取样过程对穿孔种子的发芽率、生理质量和活力没有显著影响。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

HardwareX
Engineering-Industrial and Manufacturing Engineering
CiteScore
4.10
自引率
18.20%
发文量
124
审稿时长
24 weeks
期刊介绍:
HardwareX is an open access journal established to promote free and open source designing, building and customizing of scientific infrastructure (hardware). HardwareX aims to recognize researchers for the time and effort in developing scientific infrastructure while providing end-users with sufficient information to replicate and validate the advances presented. HardwareX is open to input from all scientific, technological and medical disciplines. Scientific infrastructure will be interpreted in the broadest sense. Including hardware modifications to existing infrastructure, sensors and tools that perform measurements and other functions outside of the traditional lab setting (such as wearables, air/water quality sensors, and low cost alternatives to existing tools), and the creation of wholly new tools for either standard or novel laboratory tasks. Authors are encouraged to submit hardware developments that address all aspects of science, not only the final measurement, for example, enhancements in sample preparation and handling, user safety, and quality control. The use of distributed digital manufacturing strategies (e.g. 3-D printing) is encouraged. All designs must be submitted under an open hardware license.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


