A proteomics investigation of primary human articular chondrocyte isolation
IF 2.8
引用次数: 0
Abstract
Objective
Human primary articular chondrocytes (hPACs) are routinely isolated from articular cartilage for pre-clinical OA research. Collagenase digest of tissue is an essential step, yet the impact of lengthy enzymatic incubation on the hPAC phenotype is unclear. We aimed to delineate this through proteomic analysis.
Design
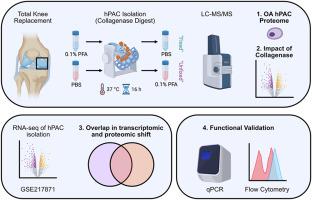
hPACs were isolated from human knee cartilage (n = 4) from patients undergoing total knee replacement. Collagenase treatment was performed with or without prior fixation of the tissue. Proteomes were quantified using LC-MS/MS. The Proteomic Ruler was employed to estimate protein copy numbers and cell protein masses. Significant differences in protein intensities were determined using paired t-testing and Benjamini-Hochberg correction. Proteomic data were integrated with existing transcriptomes (GSE217871) of hPACs and ground cartilage (ex vivo) RNA.
Results
Following collagenase treatment, we identified 498 differentially expressed proteins (DEP) in the unfixed cells. We observed depletion of FOXO signaling and enrichment of ribosomal RNA processing, indicative of increased cell cycle progression. This was supported by depletion of cell cycle inhibitors including CDKN1C. Transcriptomic analysis identified 3937 differentially expressed genes (DEG), and a 53 % overlap in DEP and DEG. Propidium iodide staining did not identify significant differences in cell cycle between fixed and unfixed hPACs.
Conclusions
We identified shifts in the proteome and transcriptome of hPACs following collagenase digest, supporting the use of tissue fixation before extracting nucleic acids for analysis where possible. Despite widespread expression changes, hPACs largely retain their chondrocyte phenotype. These datasets and analyses will serve as a valuable resource for the OA and cartilage research community.
原代人关节软骨细胞分离的蛋白质组学研究
目的:从关节软骨中常规分离人原代关节软骨细胞(hPACs)用于临床前OA研究。胶原酶消化组织是一个必要的步骤,但长时间的酶孵育对hPAC表型的影响尚不清楚。我们的目标是通过蛋白质组学分析来描述这一点。DesignhPACs从全膝关节置换术患者的膝关节软骨中分离出来(n = 4)。胶原酶治疗在事先或不事先固定组织的情况下进行。采用LC-MS/MS对蛋白质组进行定量分析。蛋白质组学尺用于估计蛋白质拷贝数和细胞蛋白质量。使用配对t检验和Benjamini-Hochberg校正来确定蛋白质强度的显著差异。蛋白质组学数据与现有的hPACs和软骨(离体)RNA转录组(GSE217871)相结合。结果经胶原酶处理后,我们在未固定细胞中鉴定出498个差异表达蛋白(DEP)。我们观察到FOXO信号的缺失和核糖体RNA加工的富集,表明细胞周期进程加快。这得到了包括CDKN1C在内的细胞周期抑制剂耗竭的支持。转录组学分析鉴定出3937个差异表达基因(DEG), DEP和DEG有53%的重叠。碘化丙啶染色未发现固定和未固定hPACs在细胞周期上有显著差异。结论:我们发现胶原酶消化后hPACs的蛋白质组和转录组发生了变化,支持在提取核酸进行分析之前使用组织固定。尽管广泛的表达改变,hPACs在很大程度上保留了其软骨细胞表型。这些数据集和分析将成为OA和软骨研究界的宝贵资源。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Osteoarthritis and cartilage open
Orthopedics, Sports Medicine and Rehabilitation
CiteScore
3.30
自引率
0.00%
发文量
0
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


