Transcriptomic and metabolomic profiling reveals key mechanisms of alkaline stress tolerance in rice
IF 2.5
4区 生物学
Q3 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Background
Alkaline stress severely restricts rice growth and yield by disrupting ion balance, nutrient uptake, and oxidative metabolism. Clarifying the molecular mechanisms of tolerance is vital for breeding resilient varieties. This study explores transcriptional and metabolic adaptations in an alkali-tolerant (Qijing 10, LD) and sensitive (Tengxi 138, WL) rice variety under alkaline stress.
Results
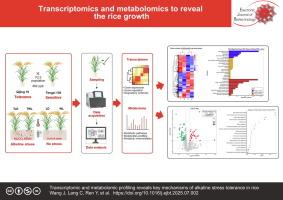
Transcriptomic analysis revealed 1297 differentially expressed genes (DEGs) in the sensitive variety under alkaline stress (TWL), primarily enriched in pathways related to antioxidant enzyme synthesis (e.g., peroxidase genes), transmembrane ion transport, and membrane lipid stabilization pathways. In contrast, the tolerant variety (TLD) exhibited only 38 DEGs, suggesting transcriptional homeostasis achieved via suppression of stress-related gene overactivation. Metabolomic profiling demonstrated stable levels of key lipids (phosphatidic acid, galactolipids) and osmolytes (proline, betaine) in the tolerant variety under stress, whereas the sensitive variety accumulated lipid peroxidation products (malondialdehyde, MDA) and displayed dysregulated carbohydrate metabolic dysregulation. Integrated multi-omics analysis indicated that the tolerant variety coordinated lipid metabolism gene modulation with antioxidant metabolite accumulation, establishing dual barriers for ROS scavenging and membrane protection. Conversely, transcriptional dysregulation in the sensitive variety led to metabolic collapse.
Conclusions
Alkaline tolerance in rice hinges on the synergistic modulation of stress-responsive genes and metabolic networks to preserve redox equilibrium and membrane function. The tolerant variety’s capacity to stabilize transcriptional activity and metabolic flux underlies its resilience. These results elucidate key molecular and metabolic determinants of alkaline tolerance, offering strategic targets for breeding rice cultivars adapted to alkaline environments.
How to cite: Wang J, Lang C, Ren Y, et al. Transcriptomic and metabolomic profiling reveals key mechanisms of alkaline stress tolerance in rice. Electron J Biotechnol 2025;78. https://doi.org/10.1016/j.ejbt.2025.07.002.
转录组学和代谢组学分析揭示了水稻耐碱性胁迫的关键机制
碱性胁迫通过破坏离子平衡、养分吸收和氧化代谢,严重制约水稻的生长和产量。阐明耐受性的分子机制对培育抗逆性品种至关重要。本研究探讨了耐碱水稻(齐粳10号,LD)和敏感水稻(腾西138号,WL)在碱性胁迫下的转录和代谢适应。结果经转录组学分析,碱胁迫(TWL)下敏感品种存在1297个差异表达基因(DEGs),主要富集于抗氧化酶合成(如过氧化物酶基因)、跨膜离子转运和膜脂稳定等相关途径。相比之下,耐受性品种(TLD)仅表现出38度,表明通过抑制应激相关基因的过度激活实现转录稳态。代谢组学分析表明,在胁迫下,耐受性品种的关键脂质(磷脂酸、半乳糖脂)和渗透物(脯氨酸、甜菜碱)水平稳定,而敏感品种的脂质过氧化产物(丙二醛、丙二醛)积累,碳水化合物代谢失调。综合多组学分析表明,耐受性品种协调脂质代谢基因调控与抗氧化代谢物积累,建立了清除ROS和膜保护的双重屏障。相反,敏感品种的转录失调导致代谢崩溃。结论水稻耐盐碱依赖于胁迫响应基因和代谢网络的协同调节,以维持氧化还原平衡和膜功能。耐受性品种稳定转录活性和代谢通量的能力是其恢复力的基础。这些结果阐明了碱性耐受性的关键分子和代谢决定因素,为培育适应碱性环境的水稻品种提供了战略目标。引用方式:王杰,郎昌,任勇,等。转录组学和代谢组学分析揭示了水稻耐碱性胁迫的关键机制。中国生物医学工程学报(英文版);2009;38。https://doi.org/10.1016/j.ejbt.2025.07.002。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Electronic Journal of Biotechnology
工程技术-生物工程与应用微生物
CiteScore
5.60
自引率
0.00%
发文量
50
审稿时长
2 months
期刊介绍:
Electronic Journal of Biotechnology is an international scientific electronic journal, which publishes papers from all areas related to Biotechnology. It covers from molecular biology and the chemistry of biological processes to aquatic and earth environmental aspects, computational applications, policy and ethical issues directly related to Biotechnology.
The journal provides an effective way to publish research and review articles and short communications, video material, animation sequences and 3D are also accepted to support and enhance articles. The articles will be examined by a scientific committee and anonymous evaluators and published every two months in HTML and PDF formats (January 15th , March 15th, May 15th, July 15th, September 15th, November 15th).
The following areas are covered in the Journal:
• Animal Biotechnology
• Biofilms
• Bioinformatics
• Biomedicine
• Biopolicies of International Cooperation
• Biosafety
• Biotechnology Industry
• Biotechnology of Human Disorders
• Chemical Engineering
• Environmental Biotechnology
• Food Biotechnology
• Marine Biotechnology
• Microbial Biotechnology
• Molecular Biology and Genetics
•Nanobiotechnology
• Omics
• Plant Biotechnology
• Process Biotechnology
• Process Chemistry and Technology
• Tissue Engineering
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


