Merging conformational landscapes in a single consensus space with FlexConsensus algorithm
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Structural heterogeneity analysis in cryogenic electron microscopy is experiencing a breakthrough in estimating more accurate, richer and interpretable conformational landscapes derived from experimental data. The emergence of new methods designed to tackle the heterogeneity challenge reflects this new paradigm, enabling users to gain a better understanding of protein dynamics. However, the question of how intrinsically different heterogeneity algorithms compare remains unsolved, which is crucial for determining the reliability, stability and correctness of the estimated conformational landscapes. Here, to overcome the previous challenge, we introduce FlexConsenus: a multi-autoencoder neural network able to learn the commonalities and differences among several conformational landscapes, enabling them to be placed in a shared consensus space with enhanced reliability. The consensus space enables the measurement of reproducibility in heterogeneity estimations, allowing users to either focus their analysis on particles with a stable estimation of their structural variability or concentrate on specific particle subsets detected by only certain methods. FlexConsensus is a multi-autoencoder-based algorithm for merging different conformational landscapes from cryogenic electron microscopy heterogeneity analysis into a common latent space for the identification of similarities and differences among various methods. This helps in the validation of estimated conformational landscape and provides tools to streamline the heterogeneity workflow.
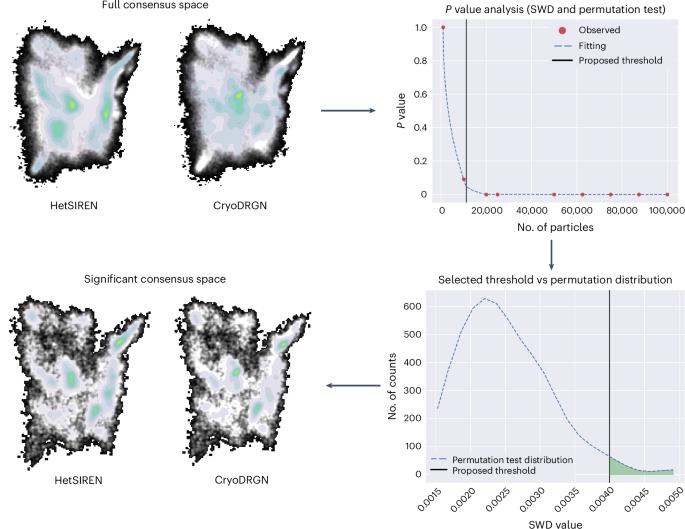
使用FlexConsensus算法合并单一共识空间中的构象景观。
低温电子显微镜的结构非均质性分析在从实验数据估计更准确、更丰富和可解释的构象景观方面取得了突破。旨在解决异质性挑战的新方法的出现反映了这种新范式,使用户能够更好地了解蛋白质动力学。然而,如何比较本质上不同的异质性算法的问题仍然没有解决,这对于确定估计的构象景观的可靠性、稳定性和正确性至关重要。在这里,为了克服之前的挑战,我们引入了flexconsensus:一个多自编码器神经网络,能够学习几个构象景观之间的共性和差异,使它们能够被放置在一个共享的共识空间中,并增强了可靠性。共识空间能够测量异质性估计的再现性,允许用户将分析集中在具有稳定估计其结构变异性的粒子上,或者集中在仅通过某些方法检测到的特定粒子子集上。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


