Structural insights into IL-31 signaling inhibition by a neutralizing antibody
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Interleukin-31 (IL-31) signals through the IL-31 receptor alpha (IL-31RA) and oncostatin M receptor beta (OSMRβ) heterodimer, mediating pruritus, dermatitis, inflammatory responses, neuroimmune interactions, and certain cancers. Here, we present the crystal structure of canine IL-31 (cIL-31) in complex with a neutralizing caninized monoclonal antibody (2D10-2). This antibody competitively inhibited cIL-31 binding to canine OSMRβ (cOSMRβ) but not to canine IL-31RA (cIL-31RA). Moreover, it effectively blocked cIL-31-induced STAT5 phosphorylation in vitro and alleviated cIL-31-induced pruritus in beagle dogs. Structural analysis identified key antibody-binding residues in α-helical A, α-helical D, and the AB loop of cIL-31. Systematic mutagenesis based on the complex structure further defined the conformational epitopes of cIL-31 recognized by cOSMRβ. In summary, this study reports the IL-31 structure, revealing a four-α-helical bundle cytokine, and elucidates 2D10-2’s neutralizing mechanism by targeting the cIL-31-cOSMRβ interaction. These findings advance our understanding of IL-31 and offer insights for developing IL-31-targeted therapeutics.
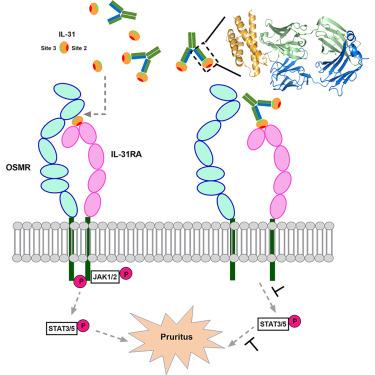
中和抗体对IL-31信号抑制的结构见解
白细胞介素-31 (IL-31)通过IL-31受体α (IL-31RA)和肿瘤抑制素M受体β (OSMRβ)异源二聚体发出信号,介导瘙痒、皮炎、炎症反应、神经免疫相互作用和某些癌症。在这里,我们展示了犬IL-31 (cIL-31)与中和犬化单克隆抗体(2D10-2)配合物的晶体结构。该抗体竞争性地抑制cIL-31与犬OSMRβ (cOSMRβ)的结合,而不抑制犬IL-31RA (cIL-31RA)的结合。体外有效阻断cil -31诱导的STAT5磷酸化,减轻了il -31诱导的beagle犬瘙痒症。结构分析鉴定了cil31的α-螺旋A、α-螺旋D和AB环的关键抗体结合残基。基于复杂结构的系统诱变进一步确定了cOSMRβ识别的cIL-31构象表位。总之,本研究报道了IL-31的结构,揭示了一个4 -α-螺旋束细胞因子,并通过靶向cIL-31-cOSMRβ相互作用阐明了2D10-2的中和机制。这些发现促进了我们对IL-31的理解,并为开发IL-31靶向治疗提供了见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


