In silico identification and field validation of diagnostic marker gene targets for the improved detection of scrub typhus
IF 1.9
4区 生物学
Q4 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Scrub typhus, a vector-borne zoonosis prevalent in the Asia-Pacific region, poses diagnostic challenges due to the pathogen's complex genome and diverse rodent and shrew hosts. The scarcity of reliable diagnostic tests hinders effective sentinel surveillance. This study aims to identify novel diagnostic gene targets using a bioinformatics approach to develop a highly sensitive and specific PCR assay for scrub typhus detection. Genome sequences of Orientia tsutsugamushi, the causative agent, were analyzed, leading to the selection of 11 potential diagnostic biomarkers. In-house conventional PCR assays targeting these biomarkers and published nested PCR assays, were tested on blood and tissue (spleen) samples of 150 field rodent and shrew. Among the tested genes, the tsa56 gene consistently demonstrated the highest detection rate in both conventional (55.6 %, n = 15) and nested (74 %, n = 20) assays, indicating it to be the most reliable diagnostic marker for scrub typhus. A novel nested PCR was designed targeting a unique tsa56 gene segment, which showed an analytical sensitivity of 4.7 (95 % CI: 2–7.4) copies/μL, and was able to detect O. tsutsugamushi in multiple hosts including human and mite samples. Moreover, fewer primer-template mismatches and no mismatches at the critical 3′ terminus with O. tsutsugamushi strains were observed. The assay did not show any cross-reaction with available non-target organisms. Thus, the developed nested PCR assay demonstrates enhanced sensitivity, specificity, and broader strain inclusivity. Overall, the study presents a promising tool for scrub typhus detection, which will aid in improved disease surveillance, outbreak prediction, and timely implementation of control measures.
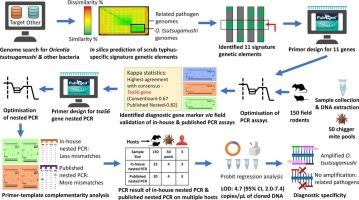
恙虫病诊断标记基因靶点的计算机鉴定及现场验证。
恙虫病是在亚太地区流行的一种媒介传播的人畜共患病,由于该病原体的复杂基因组以及啮齿动物和鼩鼱宿主的多样性,给诊断带来了挑战。缺乏可靠的诊断检测妨碍了有效的哨点监测。本研究旨在利用生物信息学方法鉴定新的诊断基因靶点,建立一种高灵敏度、高特异性的恙虫病PCR检测方法。对恙虫病病原东方体基因组序列进行分析,筛选出11个具有诊断价值的生物标志物。针对这些生物标志物的内部常规PCR分析和已发表的巢式PCR分析,在150只野外啮齿动物和鼩鼱的血液和组织(脾脏)样本上进行了测试。在检测基因中,tsa56基因在常规检测(55.6% %,n = 15)和巢式检测(74 %,n = 20)中均表现出最高的检出率,是最可靠的恙虫病诊断标记。针对tsa56基因的独特片段设计了一种新的巢式PCR,检测灵敏度为4.7(95 % CI: 2 ~ 7.4)拷贝/μL,能够检测人、螨等多种宿主的恙虫弓形虫。此外,引物模板不匹配较少,关键3′端与恙虫病体无不匹配。该试验未显示与可用的非靶生物有任何交叉反应。因此,开发的巢式PCR检测显示出更高的灵敏度、特异性和更广泛的菌株包容性。总的来说,该研究为恙虫病的检测提供了一个有前景的工具,将有助于改进疾病监测、疫情预测和及时实施控制措施。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of microbiological methods
生物-生化研究方法
CiteScore
4.30
自引率
4.50%
发文量
151
审稿时长
29 days
期刊介绍:
The Journal of Microbiological Methods publishes scholarly and original articles, notes and review articles. These articles must include novel and/or state-of-the-art methods, or significant improvements to existing methods. Novel and innovative applications of current methods that are validated and useful will also be published. JMM strives for scholarship, innovation and excellence. This demands scientific rigour, the best available methods and technologies, correctly replicated experiments/tests, the inclusion of proper controls, calibrations, and the correct statistical analysis. The presentation of the data must support the interpretation of the method/approach.
All aspects of microbiology are covered, except virology. These include agricultural microbiology, applied and environmental microbiology, bioassays, bioinformatics, biotechnology, biochemical microbiology, clinical microbiology, diagnostics, food monitoring and quality control microbiology, microbial genetics and genomics, geomicrobiology, microbiome methods regardless of habitat, high through-put sequencing methods and analysis, microbial pathogenesis and host responses, metabolomics, metagenomics, metaproteomics, microbial ecology and diversity, microbial physiology, microbial ultra-structure, microscopic and imaging methods, molecular microbiology, mycology, novel mathematical microbiology and modelling, parasitology, plant-microbe interactions, protein markers/profiles, proteomics, pyrosequencing, public health microbiology, radioisotopes applied to microbiology, robotics applied to microbiological methods,rumen microbiology, microbiological methods for space missions and extreme environments, sampling methods and samplers, soil and sediment microbiology, transcriptomics, veterinary microbiology, sero-diagnostics and typing/identification.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


