Association of As(III) Methyltransferases and Radical S-Adenosylmethionine Enzymes
IF 11.3
1区 环境科学与生态学
Q1 ENGINEERING, ENVIRONMENTAL
引用次数: 0
Abstract
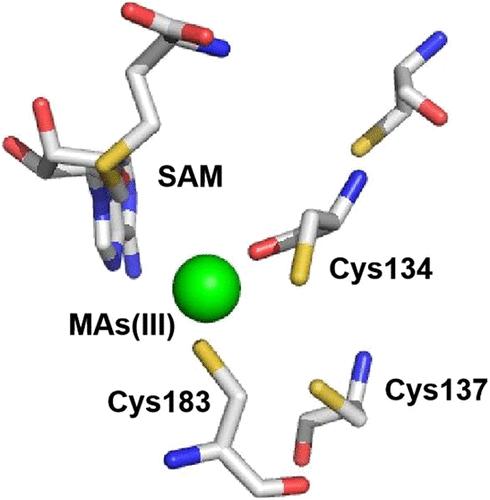
Microbial arsenic methylation catalyzed by the enzyme arsenite (As(III)) S-adenosylmethionine methyltransferase (ArsM) plays a pivotal role in the arsenic biogeochemical cycle. This produces methylated species and, when coupled with radical SAM (rSAM) enzymes, produces more complex compounds such as arsenosugars and the antibiotic arsinothricin (AST). In this study, we identified an arsM from Chloracidobacterium thermophilum that encodes the enzyme CtArsM and is adjacent to a gene for an rSAM enzyme. Most ArsMs have three domains for SAM binding, As(III) binding, and a C-terminal domain of uncertain function. CtArsM has only the first two domains. CtArsM methylates methylarsenite (MAs(III)) but not As(III) and confers resistance to MAs(III) when expressed in Escherichia coli. At 45 °C, CtArsM rapidly converts 95% of MAs(III) into volatile trimethylarsine (TMAs(III)). Mutational analysis indicates that conserved cysteine residues Cys134 and Cys183 form the MAs(III) binding site, while Cys137 is not required but increases the amount of volatilized arsenic. Mutation of Cys137 shifts the product profile, primarily resulting in dimethylarsinic acid (DMAs(V)). These findings suggest that cysteine modification can modulate ArsM activity, providing a basis for potential applications in arsenic bioremediation. Moreover, ArsM enzymes adjacent to rSAM genes may act synergistically in the biosynthesis of novel organoarsenical antibiotics.
As(III)甲基转移酶与s -腺苷甲硫氨酸自由基酶的关联
亚砷酸盐(As(III)) s -腺苷蛋氨酸甲基转移酶(ArsM)催化的微生物砷甲基化在砷的生物地球化学循环中起着关键作用。这产生甲基化的物种,当与自由基SAM (rSAM)酶结合时,产生更复杂的化合物,如砷糖和抗生素arsinthricin (AST)。在这项研究中,我们从嗜热氯酸杆菌中鉴定了一个编码CtArsM酶的arsM,它与rSAM酶的基因相邻。大多数arsm有三个SAM结合域、As(III)结合域和一个功能不确定的c端结构域。CtArsM只有前两个域。CtArsM在大肠杆菌中表达时,甲基化甲基拉森酸(MAs(III))但不甲基化As(III),并对MAs(III)具有抗性。在45°C时,CtArsM迅速将95%的MAs(III)转化为挥发性三甲基larsin (TMAs(III))。突变分析表明,保守的半胱氨酸残基Cys134和Cys183形成了MAs(III)结合位点,而Cys137不需要,但增加了砷的挥发量。Cys137的突变改变了产物谱,主要导致二甲基larsinic酸(DMAs(V))。这些结果表明,半胱氨酸修饰可以调节ArsM活性,为砷生物修复的潜在应用奠定了基础。此外,与rSAM基因相邻的ArsM酶可能在新型有机含砷抗生素的生物合成中协同作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

环境科学与技术
环境科学-工程:环境
CiteScore
17.50
自引率
9.60%
发文量
12359
审稿时长
2.8 months
期刊介绍:
Environmental Science & Technology (ES&T) is a co-sponsored academic and technical magazine by the Hubei Provincial Environmental Protection Bureau and the Hubei Provincial Academy of Environmental Sciences.
Environmental Science & Technology (ES&T) holds the status of Chinese core journals, scientific papers source journals of China, Chinese Science Citation Database source journals, and Chinese Academic Journal Comprehensive Evaluation Database source journals. This publication focuses on the academic field of environmental protection, featuring articles related to environmental protection and technical advancements.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


