DIATAGeR: Triacylglycerol annotation of data-independent acquisition based lipidomics
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
Triacylglycerols (TGs) are the most abundant lipids in the human body and the primary source of energy storage. TGs are comprised of three fatty acyls with various lengths and double bond composition, complicating structural annotation when performing lipidomics by LCMS. Data-independent acquisition (DIA) based lipidomics enables a continuous and unbiased acquisition of all TGs, creating the potential for more comprehensive TG analysis. However, TG identification in DIA lipidomics data is challenging due to the difficulty analyzing multiplexed tandem mass spectra (MS2).
Results
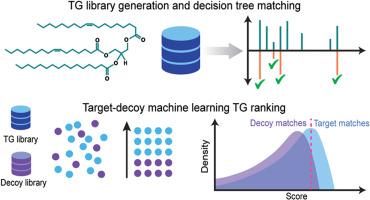
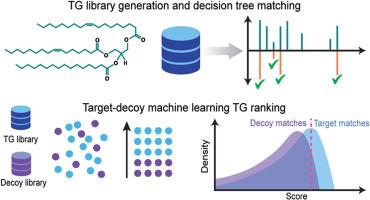
In this study, we present DIATAGeR, an R package aimed to improve and automate TG identifications to the molecular species level in DIA-based lipidomics. With DIATAGeR, TGs are identified using a TG-centric approach, where each TG in the reference database is considered as an analysis target, searched in DIA spectra, and scored using a logistic regression machine learning algorithm. Additionally, DIATAGeR uses a false discovery rate (FDR) correction calculated by a target-decoy approach to improve the confidence of TG identification and limit false positives due to interference from unrelated ions. The performance of DIATAGeR was validated in a lipidomic study of liver and plasma samples from mice with metabolic dysfunction-associated steatohepatitis (MASH) and healthy controls. All 9 TG standards were annotated at an FDR <0.1 in both datasets. When benchmarked against MS-DIAL, TGs identified by DIATAGeR contained 18 % and 12 % more even-carbon fatty acyls in liver and plasma datasets, respectively.
Significance
DIATAGeR is a valuable tool for streamlining complex TG annotation in DIA-lipidomics data. It supports vendor-neutral MS spectra data formats and offers a customizable reference database. By combining TG-centric and target-decoy approaches, DIATAGeR showed improvements in TG identification by addressing primary challenges associated with multiplexed MS2 spectra. DIATAGeR is freely available at https://github.com/Velenosi-Lab/DIATAGeR.
DIATAGeR:基于数据独立获取的脂质组学的三酰基甘油注释
甘油三酯(TGs)是人体中含量最丰富的脂类,也是能量储存的主要来源。tg由三个不同长度和双键组成的脂肪酰基组成,使LCMS进行脂质组学时的结构注释变得复杂。基于数据独立采集(DIA)的脂质组学能够连续、公正地采集所有TG,为更全面的TG分析创造了潜力。然而,由于难以分析多路串联质谱(MS2),在DIA脂质组学数据中进行TG鉴定具有挑战性。在这项研究中,我们提出了DIATAGeR,一个R包,旨在提高和自动化TG鉴定到分子物种水平的基于DIATAGeR的脂质组学。使用DIATAGeR,可以使用以TG为中心的方法识别TG,其中参考数据库中的每个TG都被视为分析目标,在DIA光谱中进行搜索,并使用逻辑回归机器学习算法进行评分。此外,DIATAGeR使用通过目标诱饵方法计算的错误发现率(FDR)校正来提高TG鉴定的置信度,并限制由于不相关离子干扰而产生的假阳性。DIATAGeR的性能在代谢功能障碍相关脂肪性肝炎(MASH)小鼠和健康对照小鼠的肝脏和血浆样本的脂质组学研究中得到了验证。在两个数据集中,所有9个TG标准的FDR均为0.1。当以MS-DIAL为基准时,DIATAGeR鉴定的tg在肝脏和血浆数据集中分别含有18%和12%的均匀碳脂肪酰基。significance ediatager是一种有价值的工具,用于简化dia -脂质组学数据中复杂的TG注释。它支持供应商中立的质谱数据格式,并提供可定制的参考数据库。通过结合以TG为中心和目标诱饵的方法,DIATAGeR通过解决与多路MS2光谱相关的主要挑战,改善了TG识别。DIATAGeR可在https://github.com/Velenosi-Lab/DIATAGeR免费获得。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


