Modelling and design of transcriptional enhancers
IF 37.6
引用次数: 0
Abstract
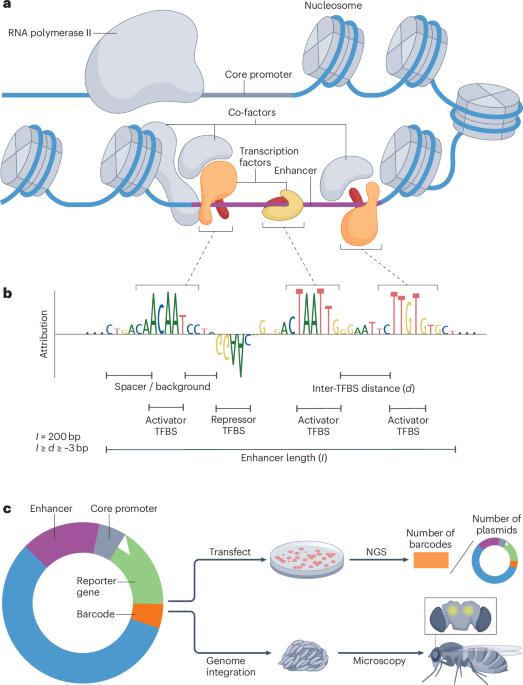
Transcriptional enhancers are the genomic elements that contain critical information for the regulation of gene expression. This information is encoded through precisely arranged transcription factor-binding sites. Genomic sequence-to-function models, computational models that take DNA sequences as input and predict gene regulatory features, have become essential for unravelling the complex combinatorial rules that govern cell-type-specific activities of enhancers. These models function as biological ‘oracles’, capable of accurately predicting the activity of novel DNA sequences. By leveraging these oracles, DNA sequences can be optimized towards designed synthetic enhancers with tailored cell-type-specific or cell-state-specific activities. In parallel, generative artificial intelligence is rapidly advancing in genomics and enhancer design. Synthetic enhancers hold great promise for a wide range of biomedical applications, from facilitating fundamental research to enabling gene therapies. Enhancers are genomic elements critical for regulating gene expression. In this Review, the authors discuss how sequence-to-function models can be used to unravel the rules underlying enhancer activity and function as biological ‘oracles’ aiding the design of synthetic enhancers with tailored cell-type-specific or cell-state-specific activities.
转录增强子的建模和设计
转录增强子是包含基因表达调控关键信息的基因组元件。这些信息是通过精确排列的转录因子结合位点编码的。基因组序列-功能模型,即以DNA序列为输入并预测基因调控特征的计算模型,已经成为揭示控制增强子细胞类型特异性活动的复杂组合规则的关键。这些模型就像生物学上的“预言器”,能够准确地预测新DNA序列的活性。通过利用这些神谕,DNA序列可以优化为具有量身定制的细胞类型特异性或细胞状态特异性活性的设计合成增强子。与此同时,生成式人工智能在基因组学和增强子设计方面也在迅速发展。从促进基础研究到实现基因治疗,合成增强剂在广泛的生物医学应用中具有很大的前景。增强子是调控基因表达的关键基因组元件。在这篇综述中,作者讨论了如何使用序列-功能模型来揭示增强子活性和功能的潜在规则,作为生物学“预言器”,帮助设计具有量身定制的细胞类型特异性或细胞状态特异性活性的合成增强子。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


